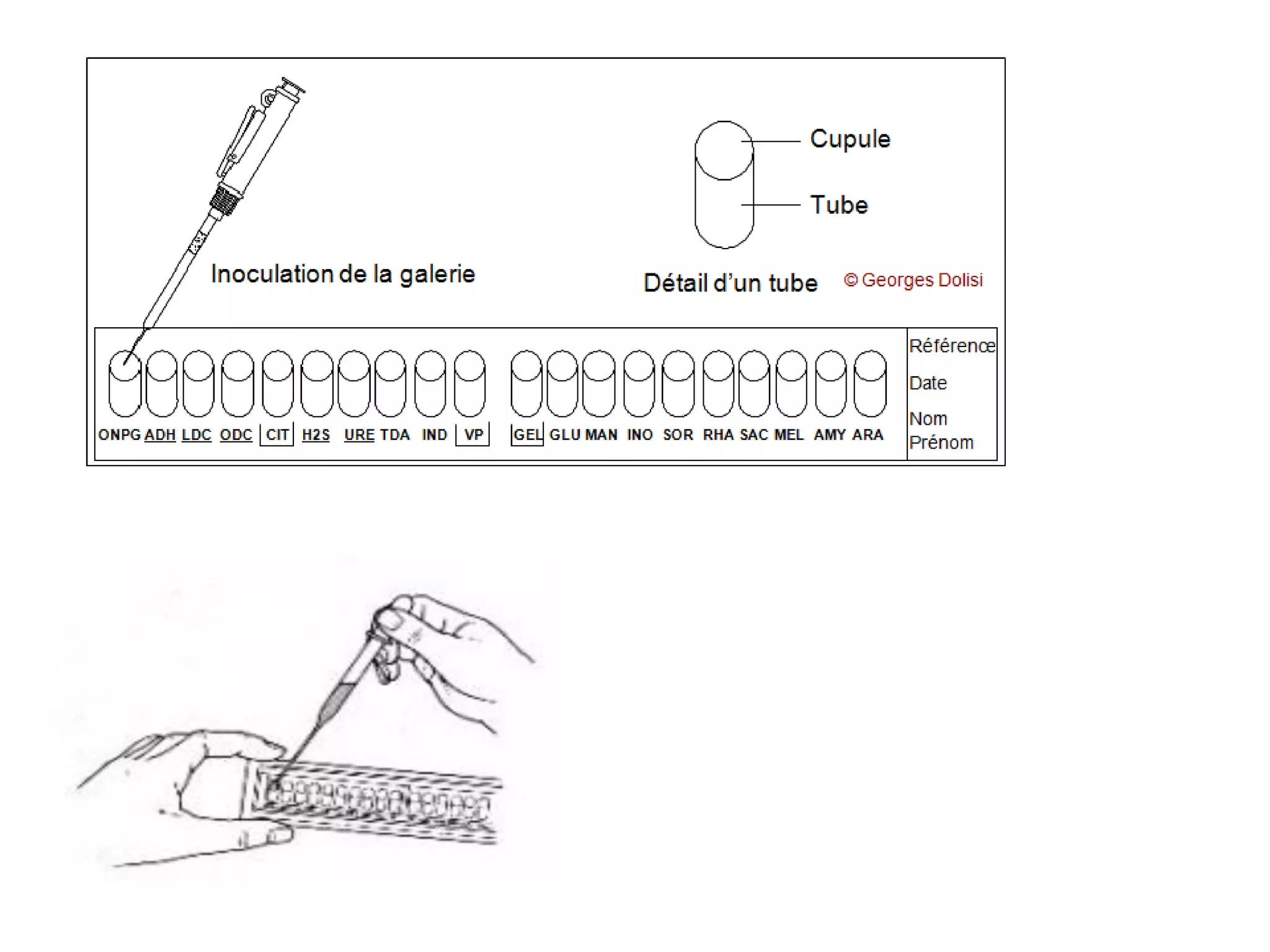
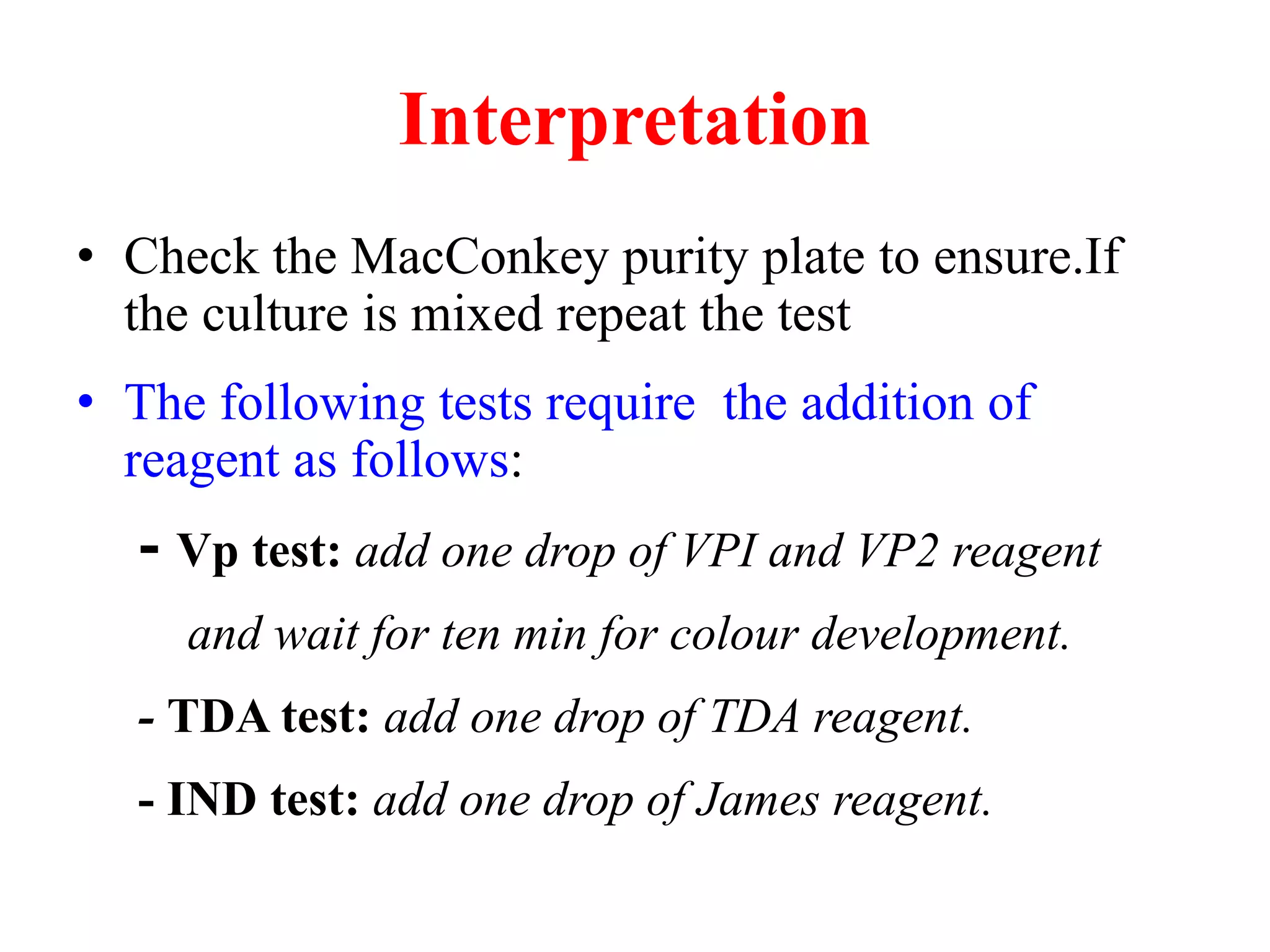
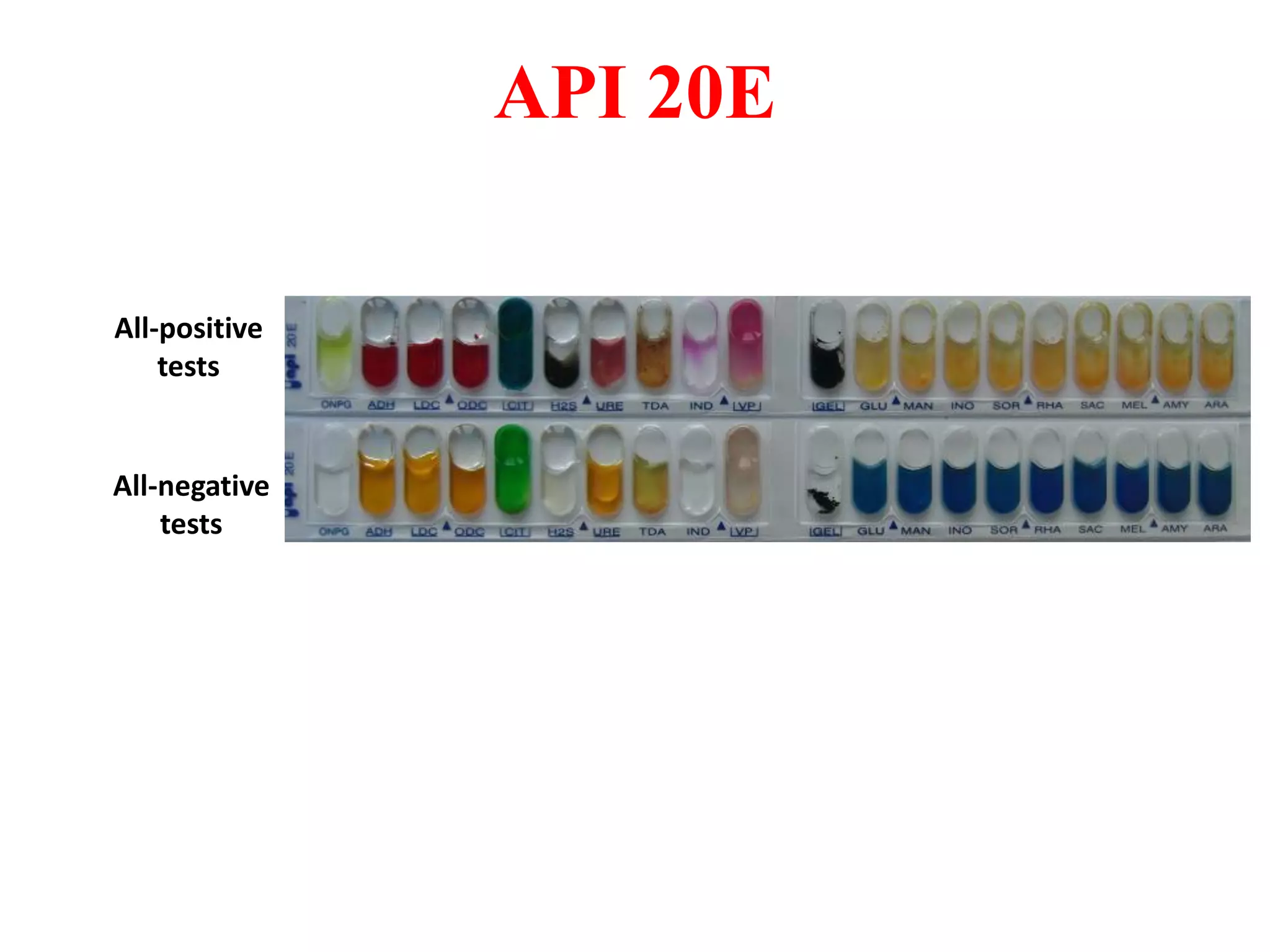
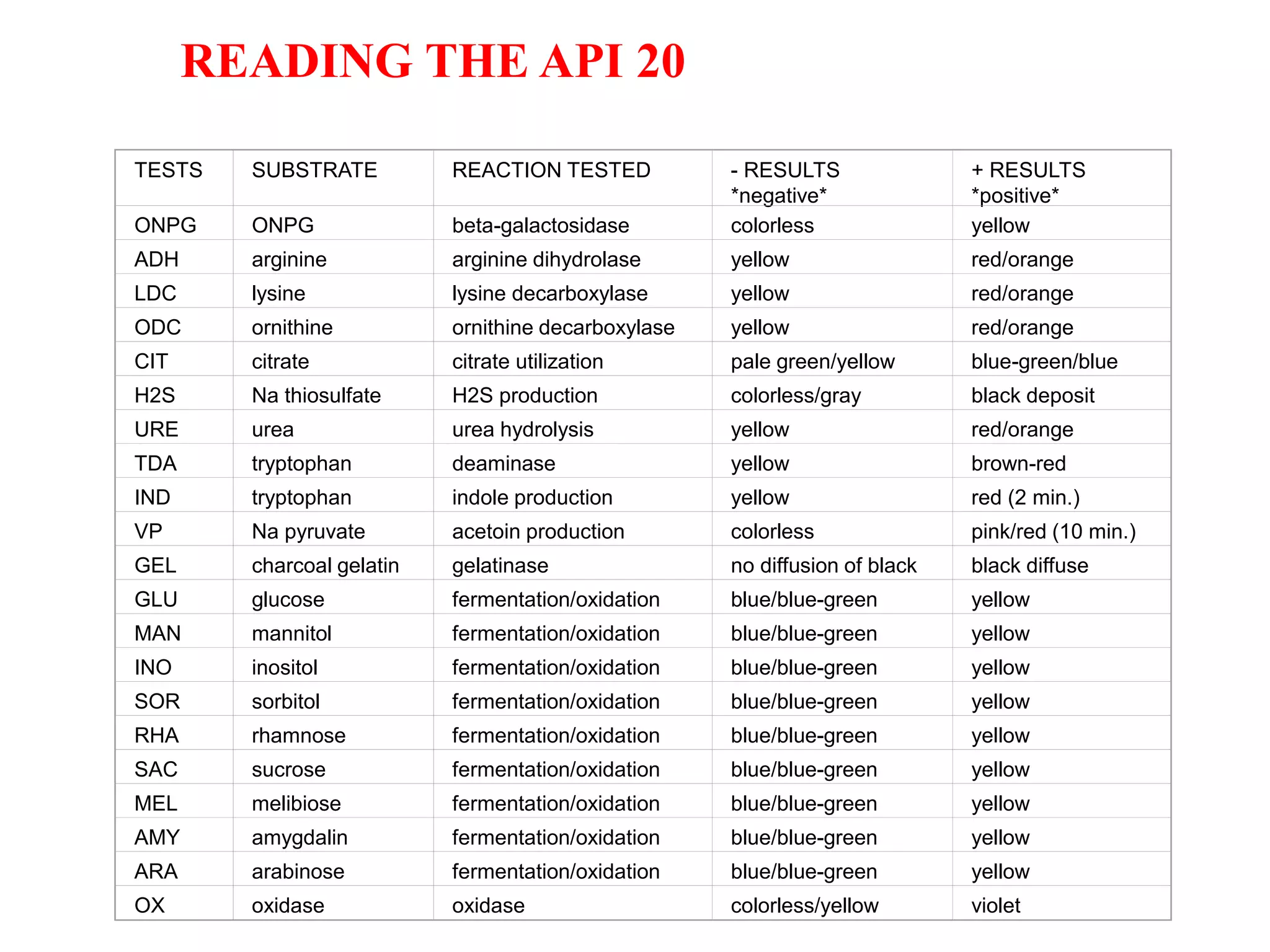
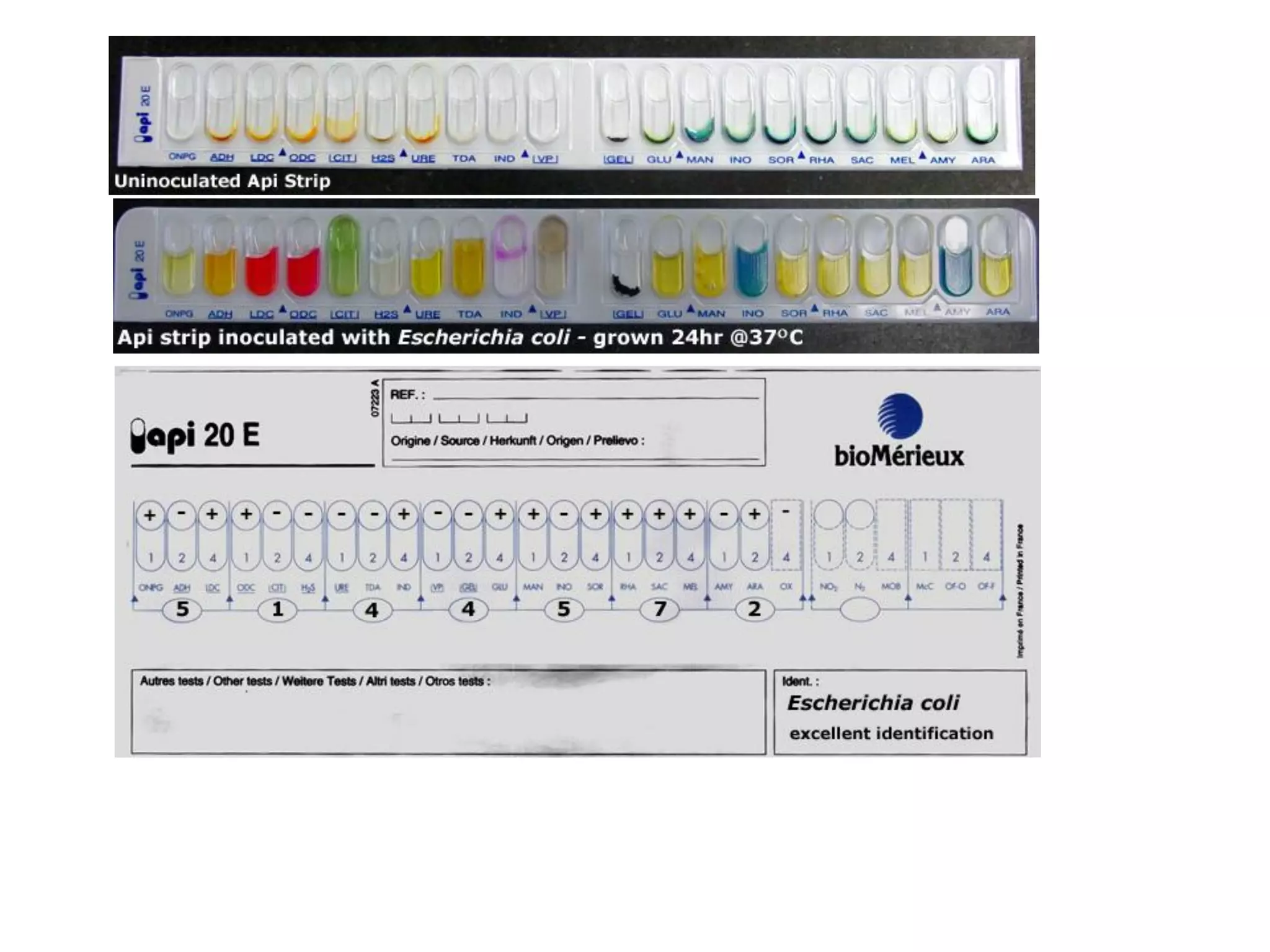
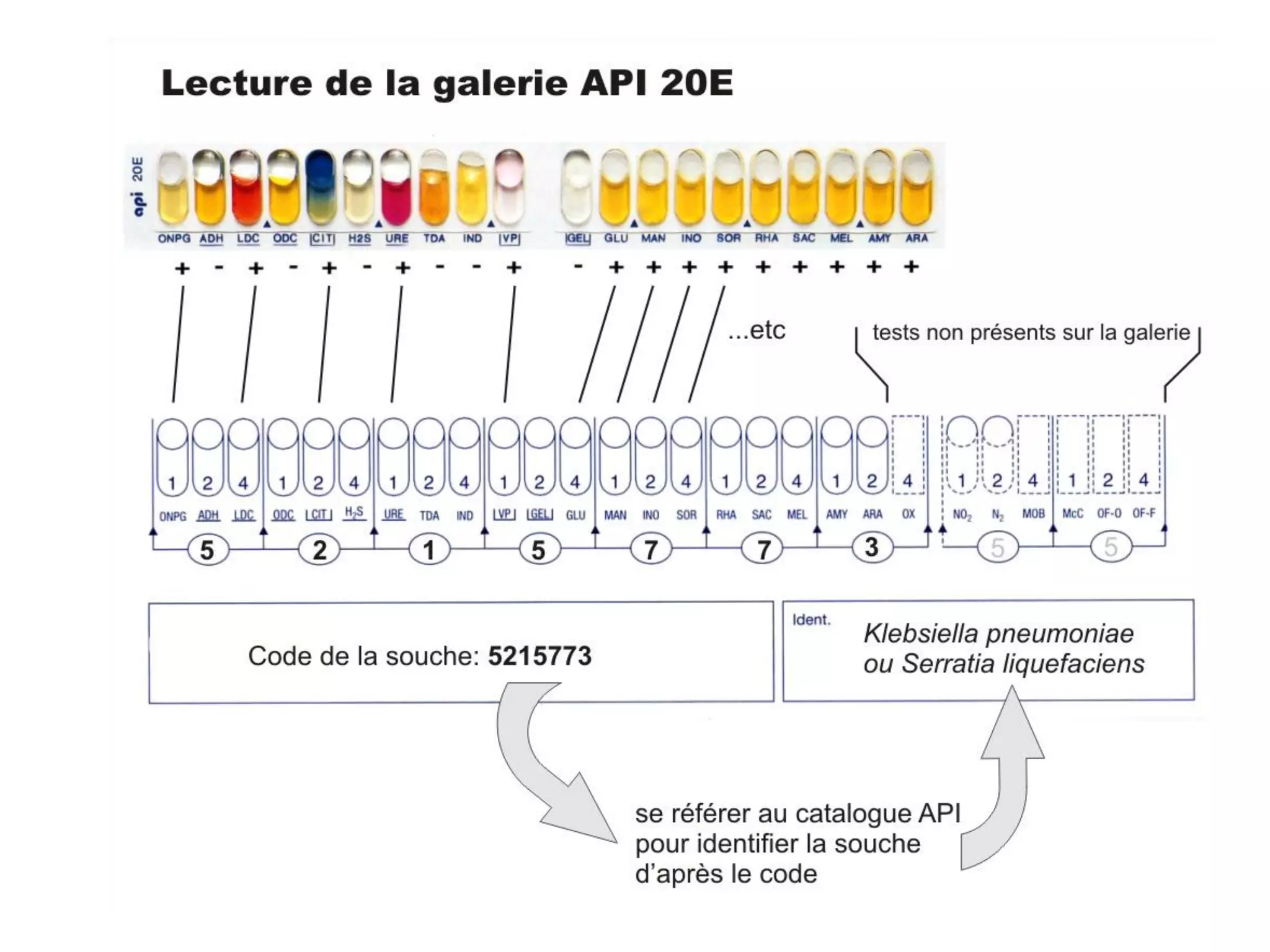
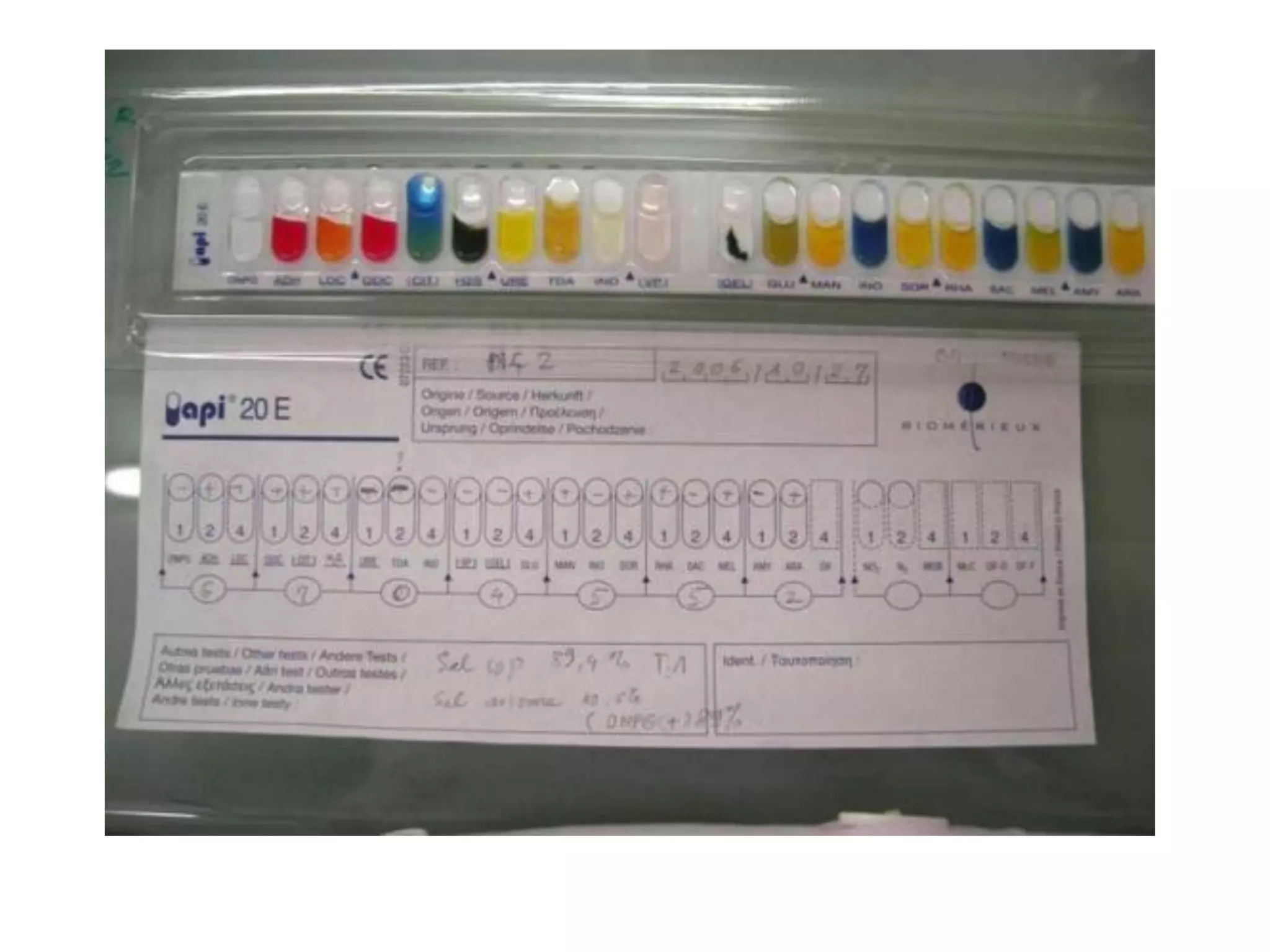
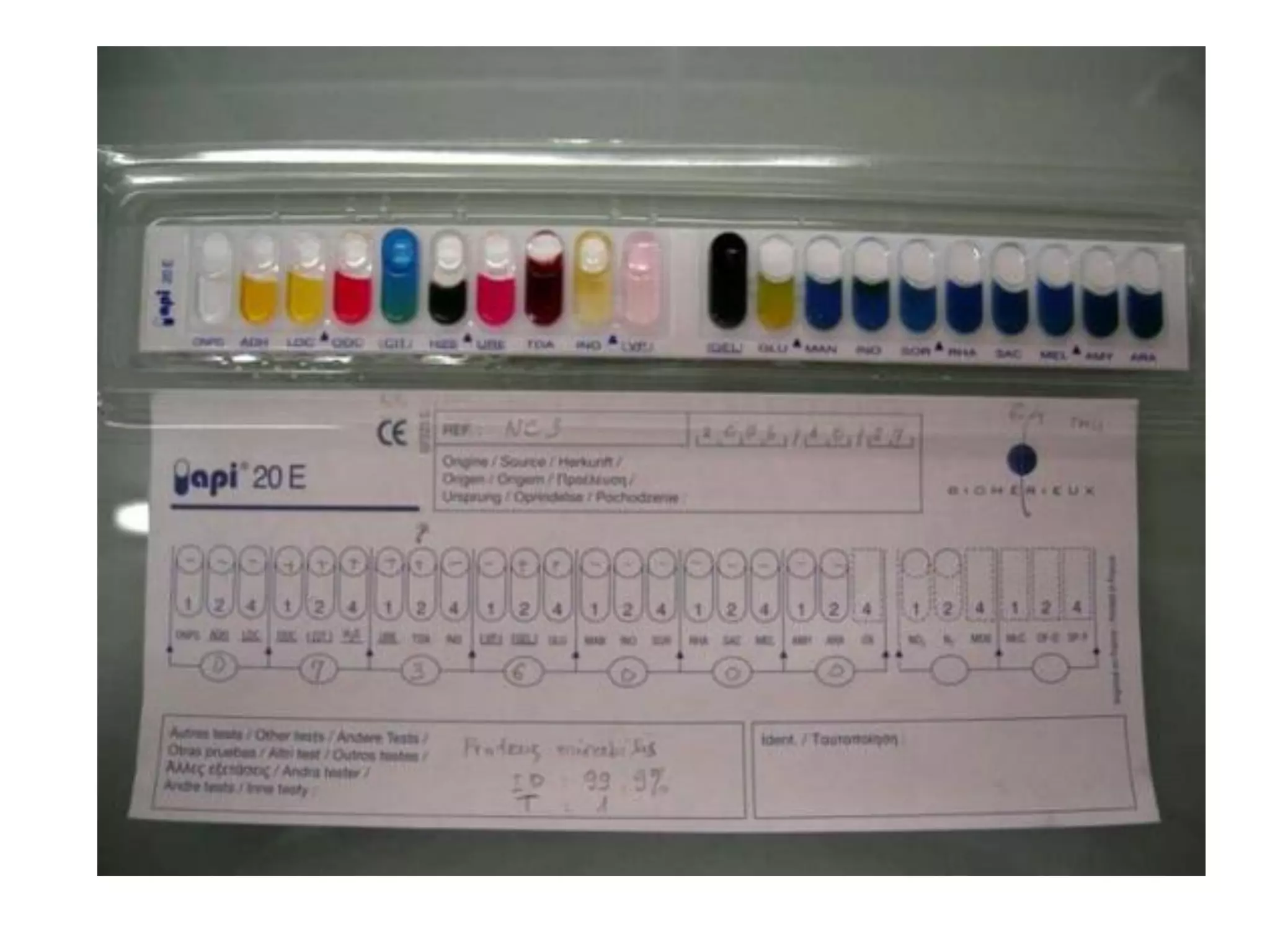
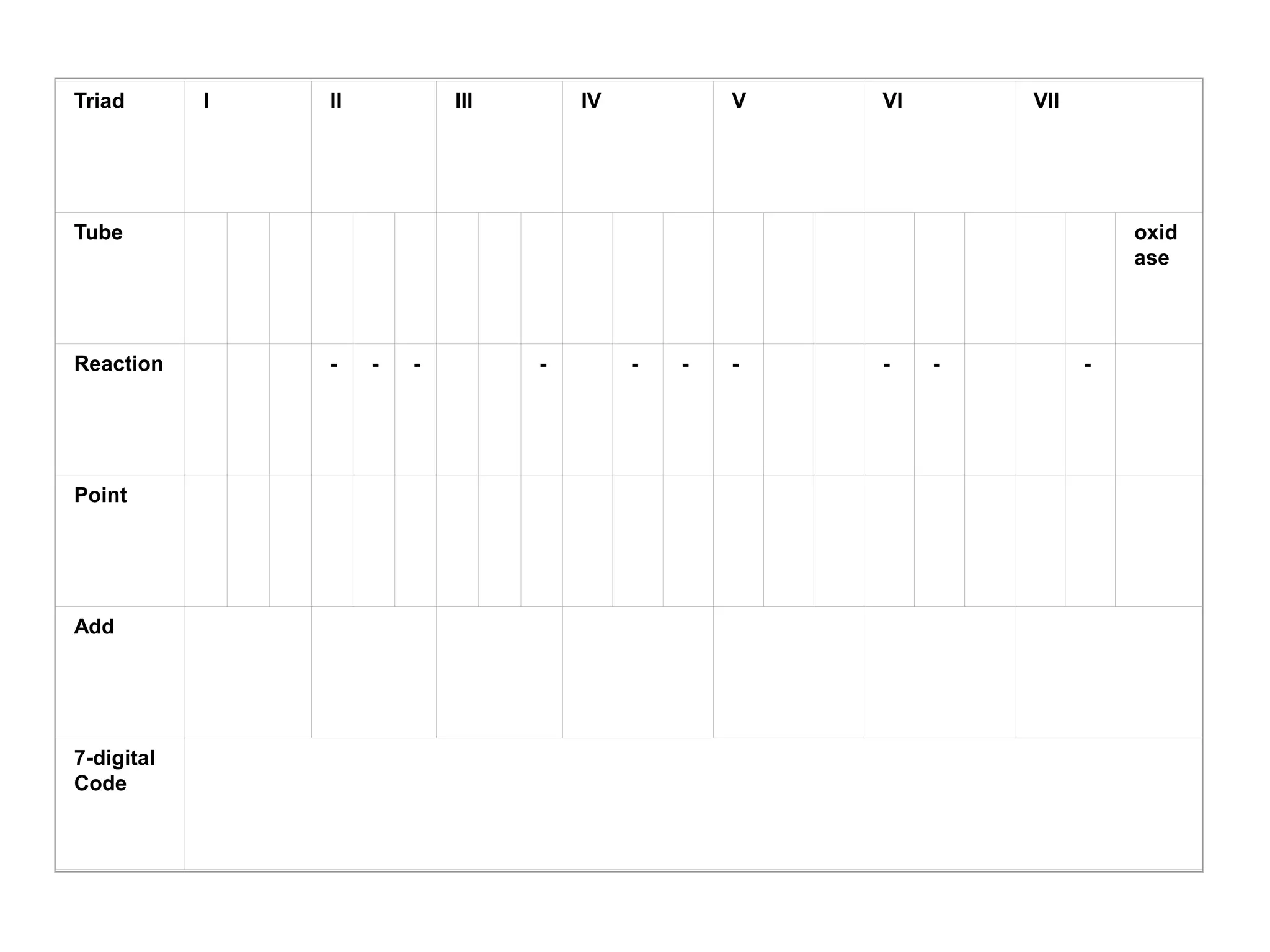
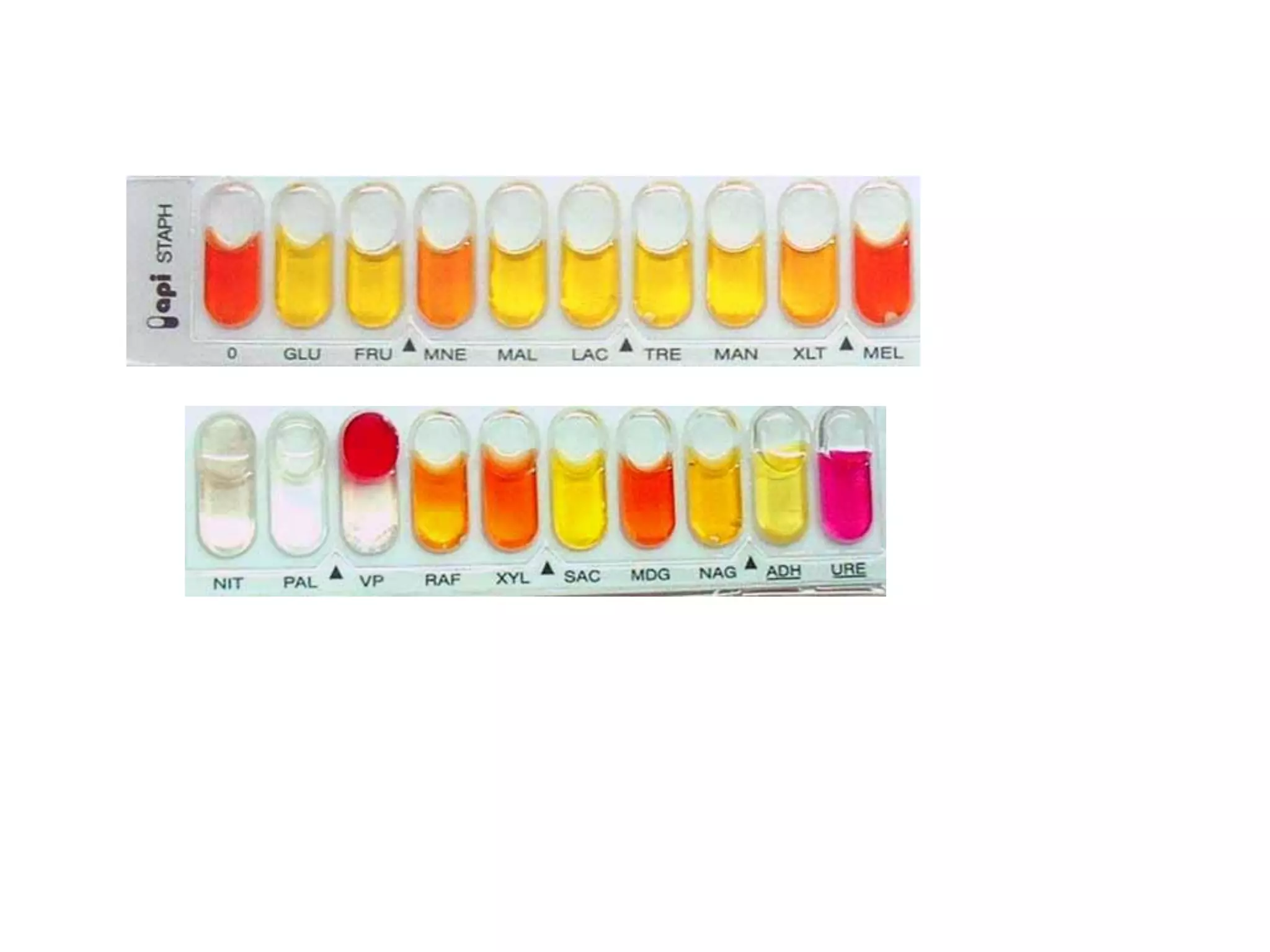
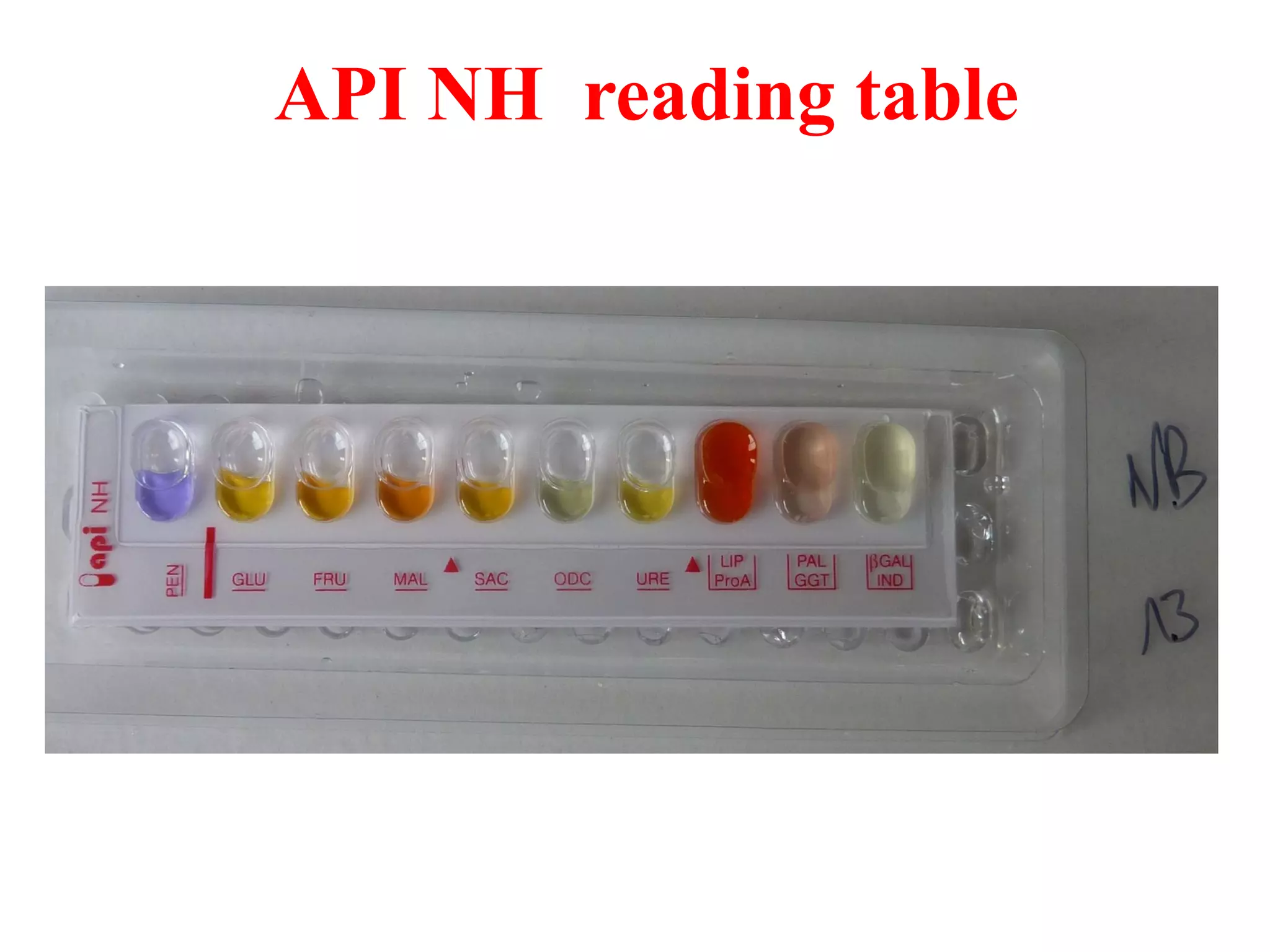
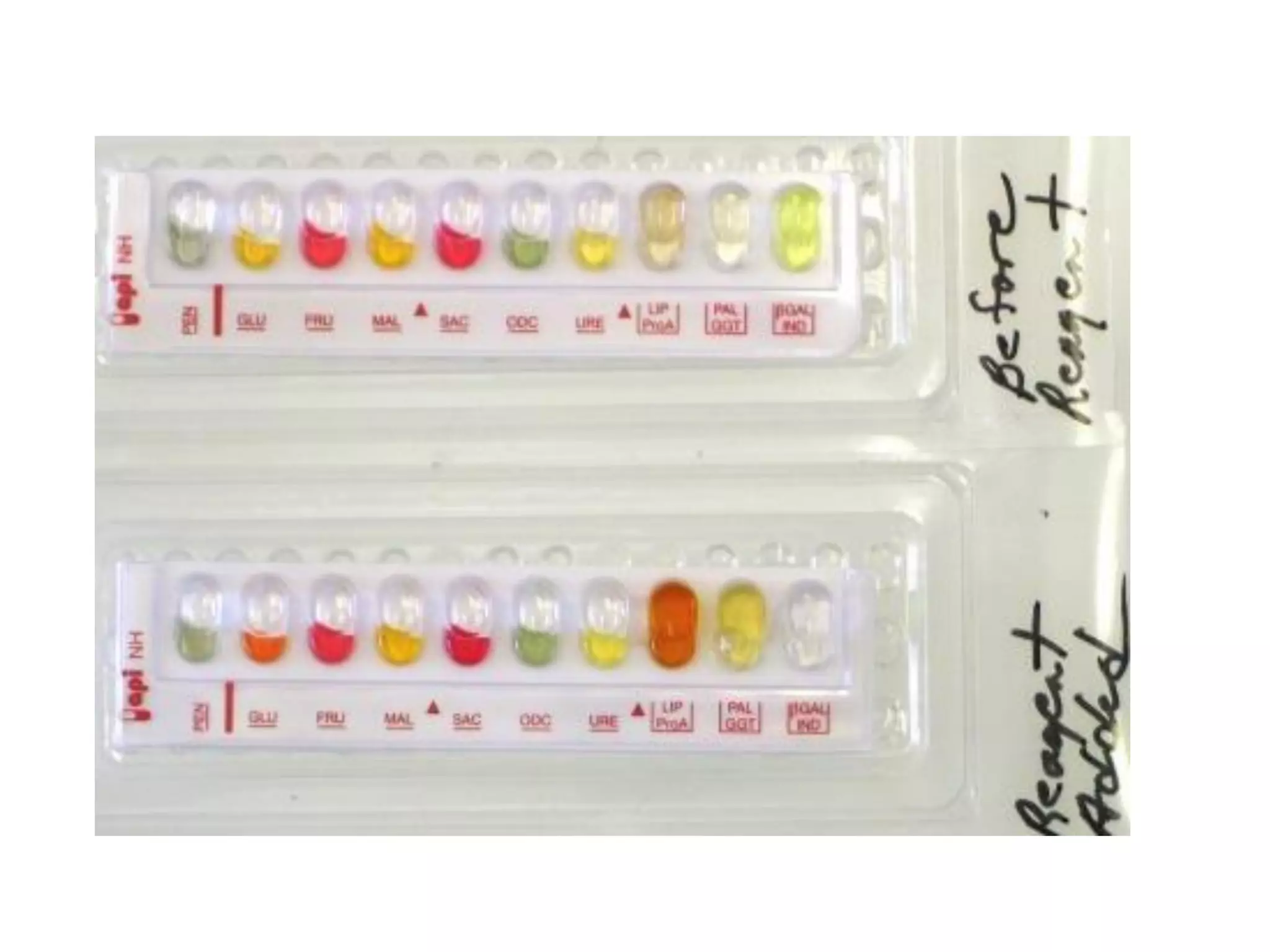
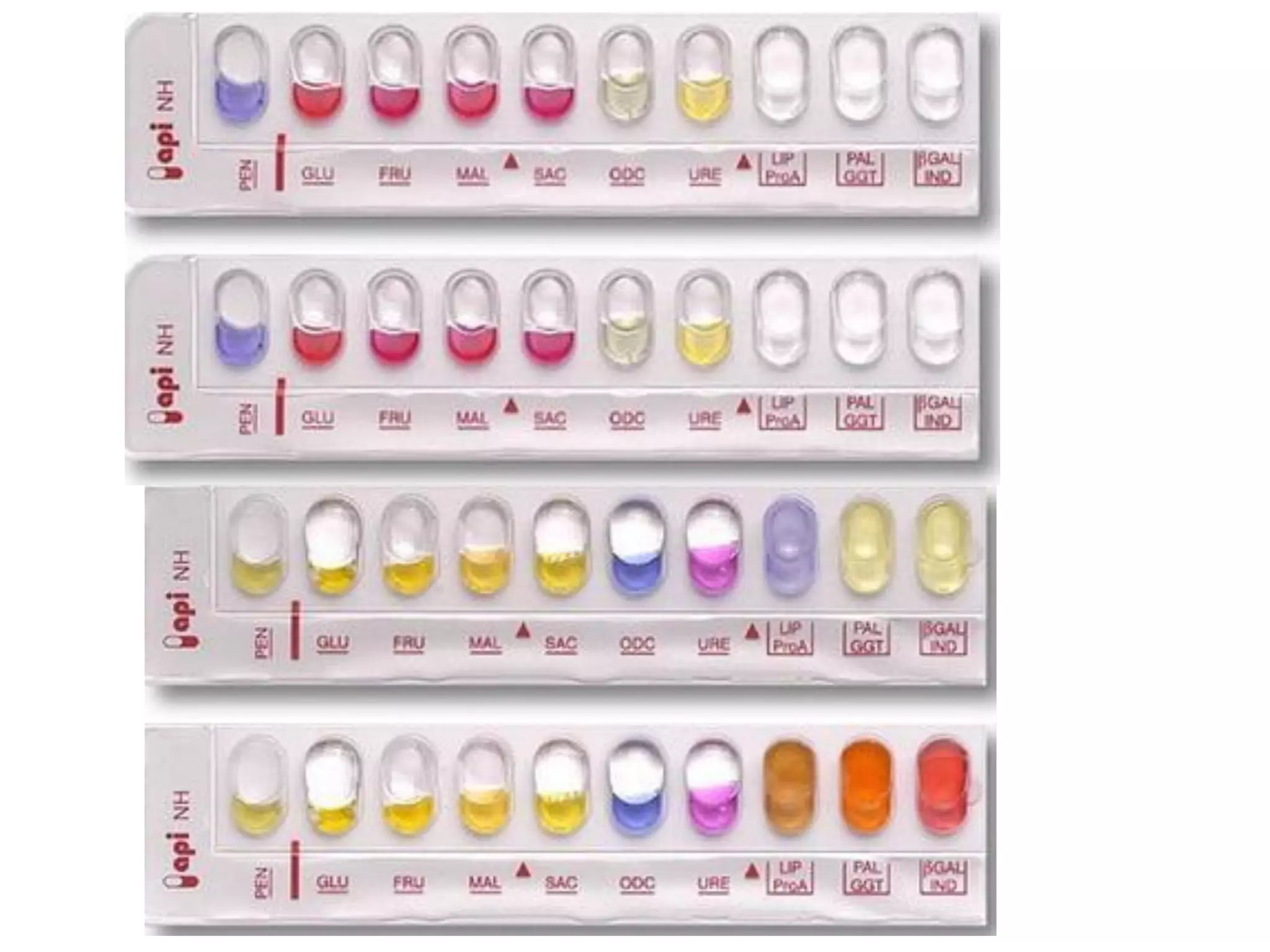
The document discusses the API (analytical profile index) system for bacterial identification. The API system uses biochemical tests contained in strips to generate identification profiles for bacteria. Different API strips exist for identifying gram-negative bacteria (API 20E), gram-positive bacteria (API 20 Strep, API 20 Staph), and anaerobes (API 20A). The API 20E strip contains 20 microtubes with dehydrated substrates that are inoculated to identify Enterobacteriaceae and other gram-negatives through color change reactions over 18-24 hours.