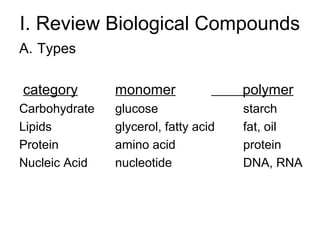
1) The document reviews biological compounds including carbohydrates, lipids, proteins, and nucleic acids.
2) It describes the structures of proteins including primary, secondary, tertiary, and quaternary structure.
3) Nucleic acids are composed of nucleotides that are made up of phosphate groups, sugars (ribose or deoxyribose), and nitrogenous bases. DNA and RNA differ in their sugar and base components.