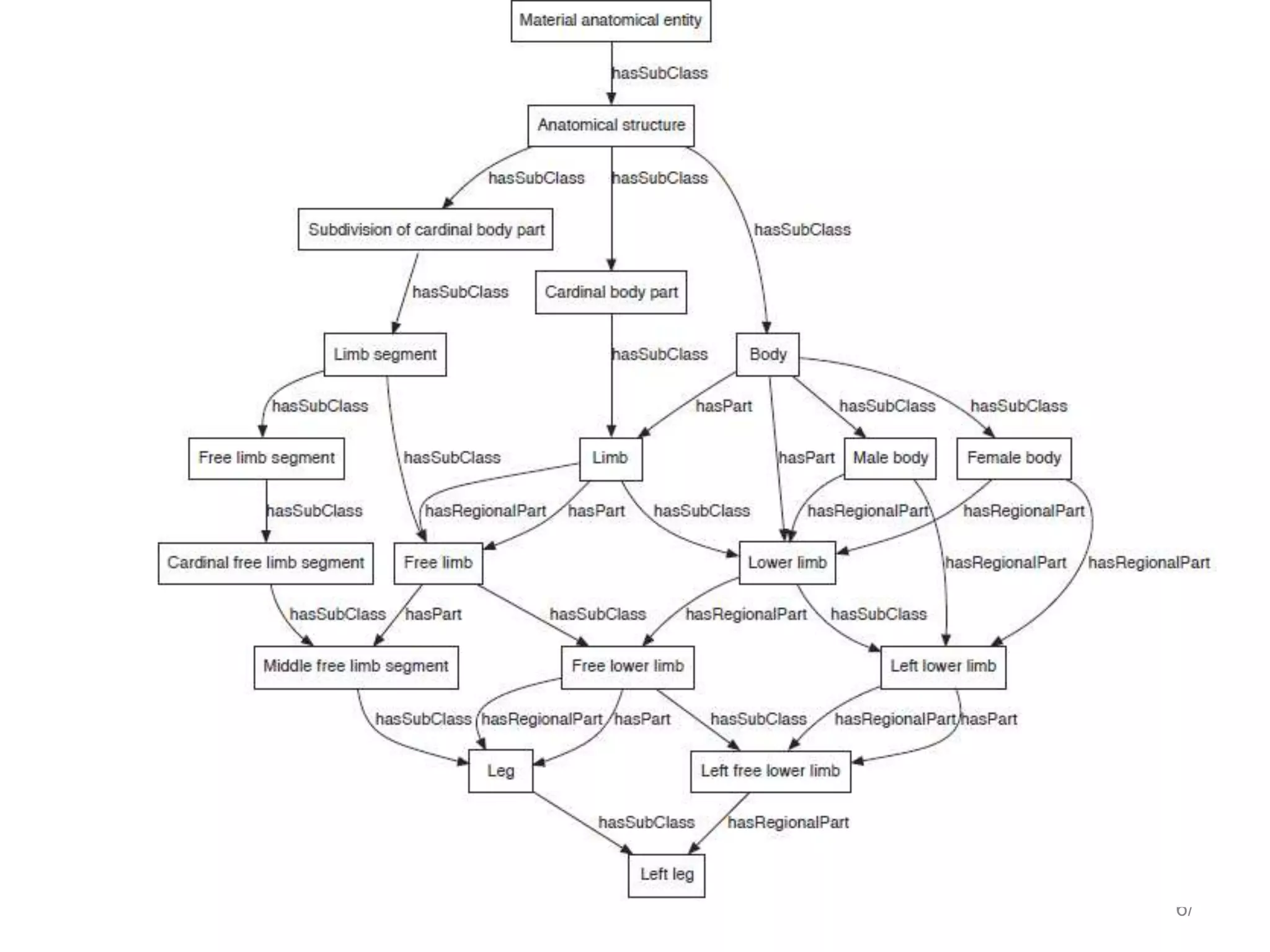
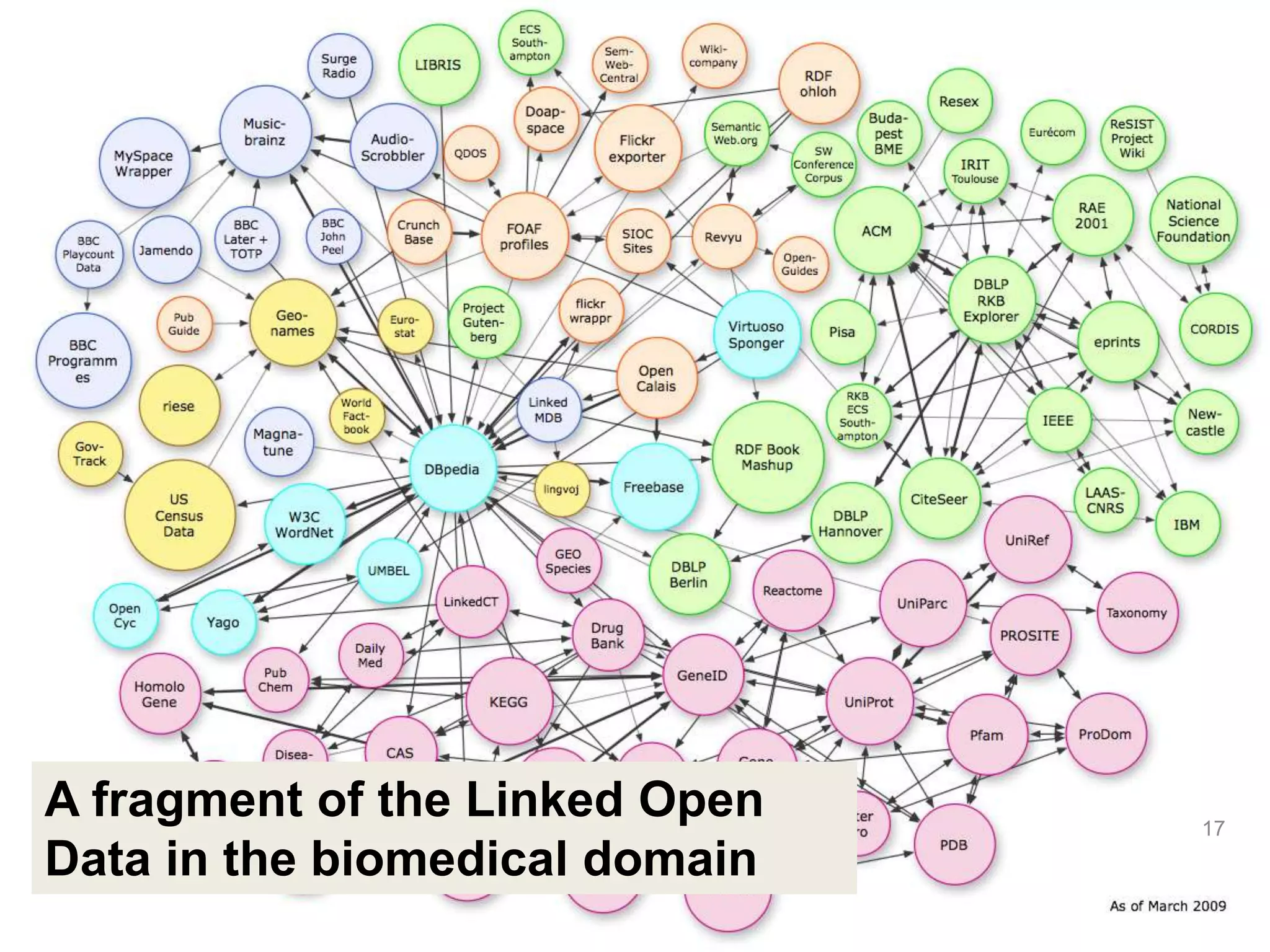
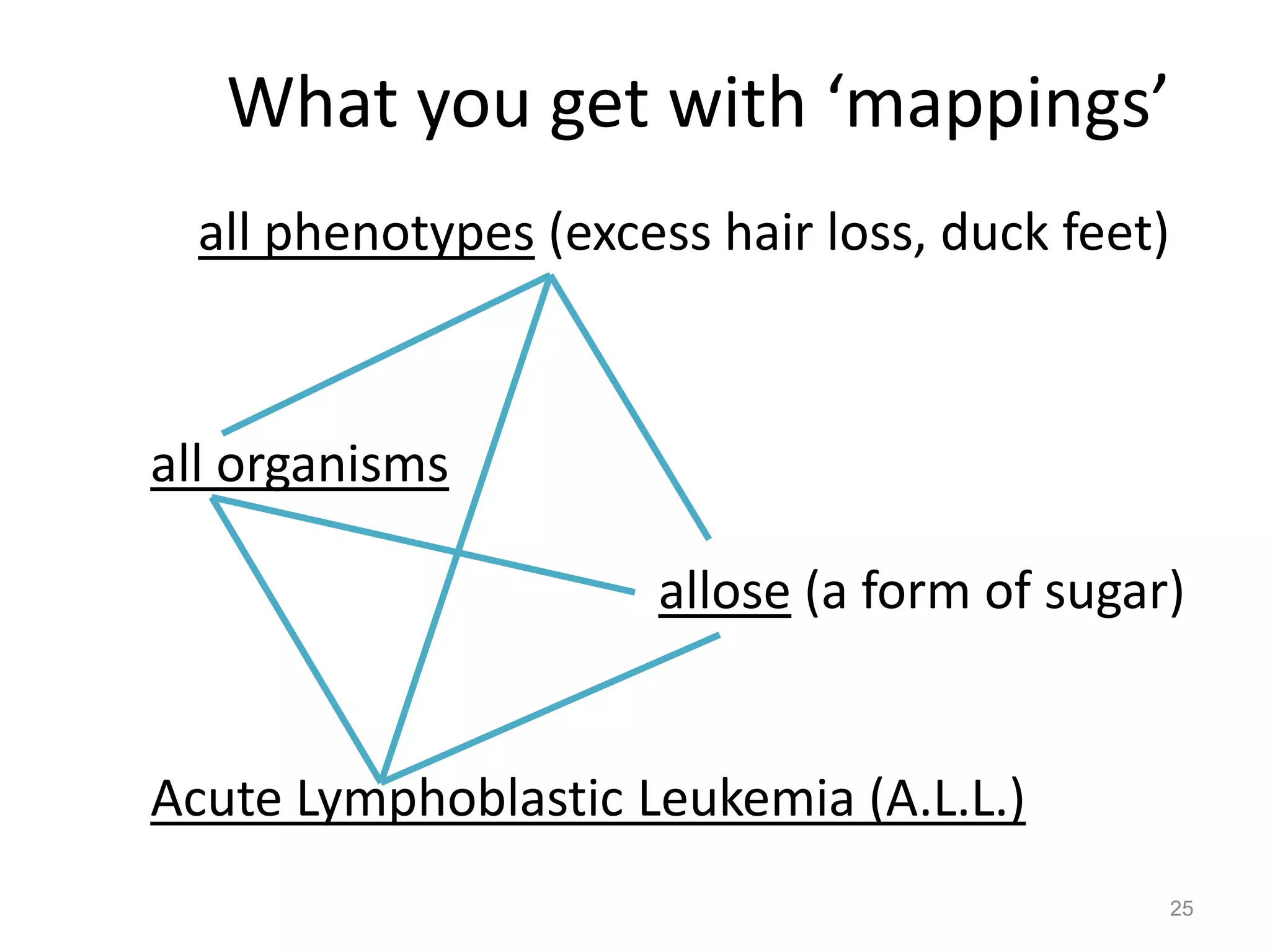
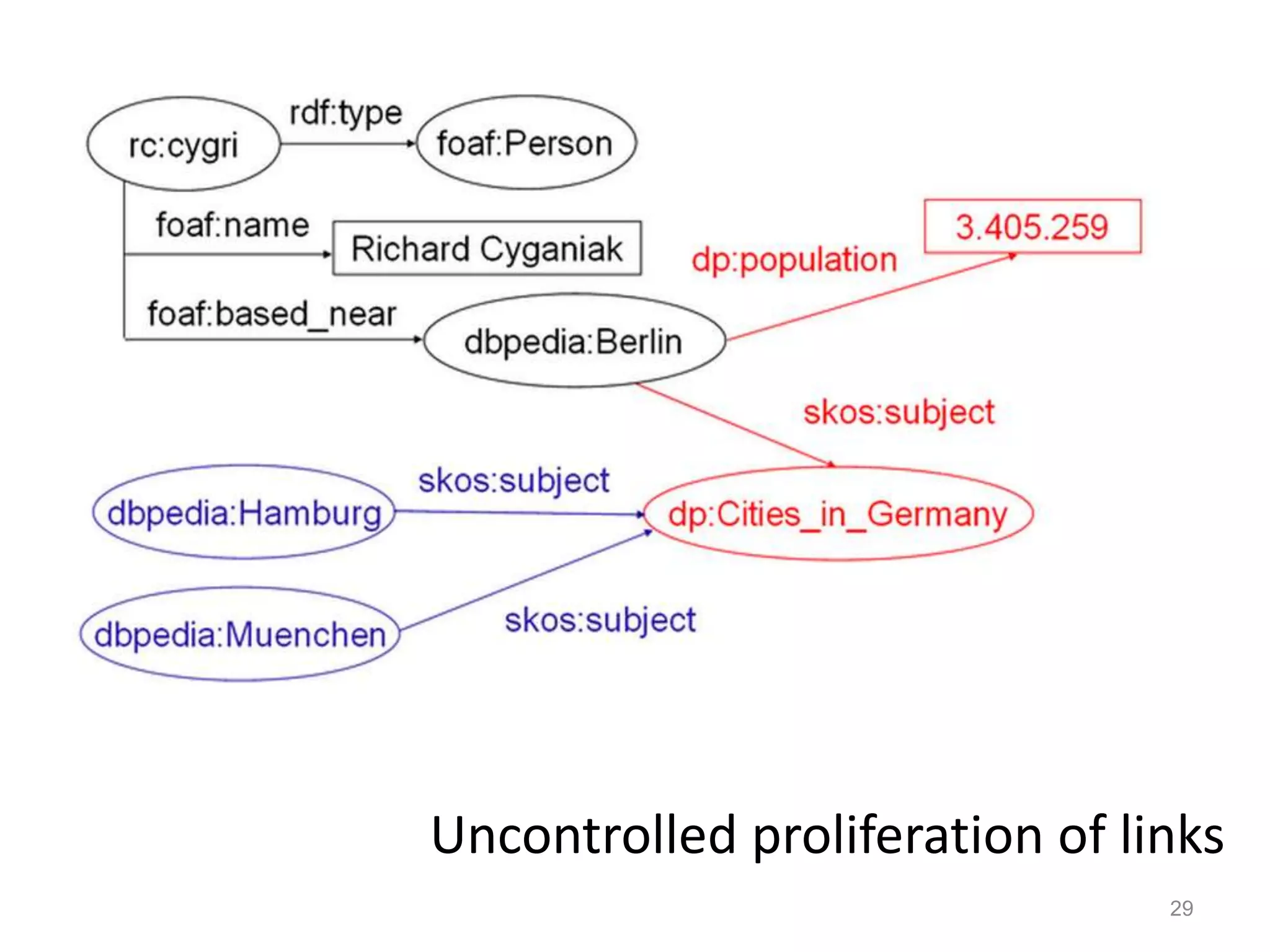
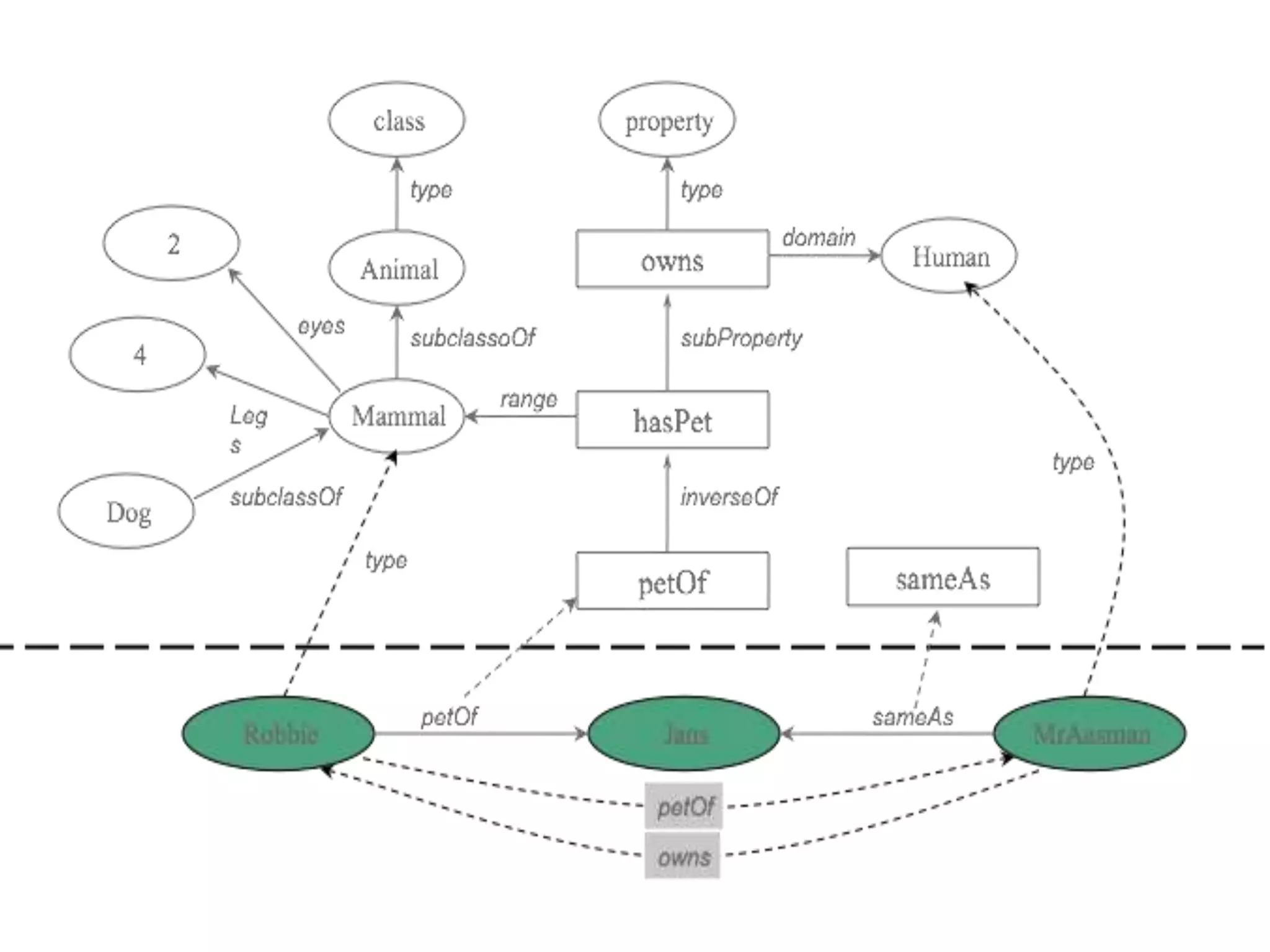
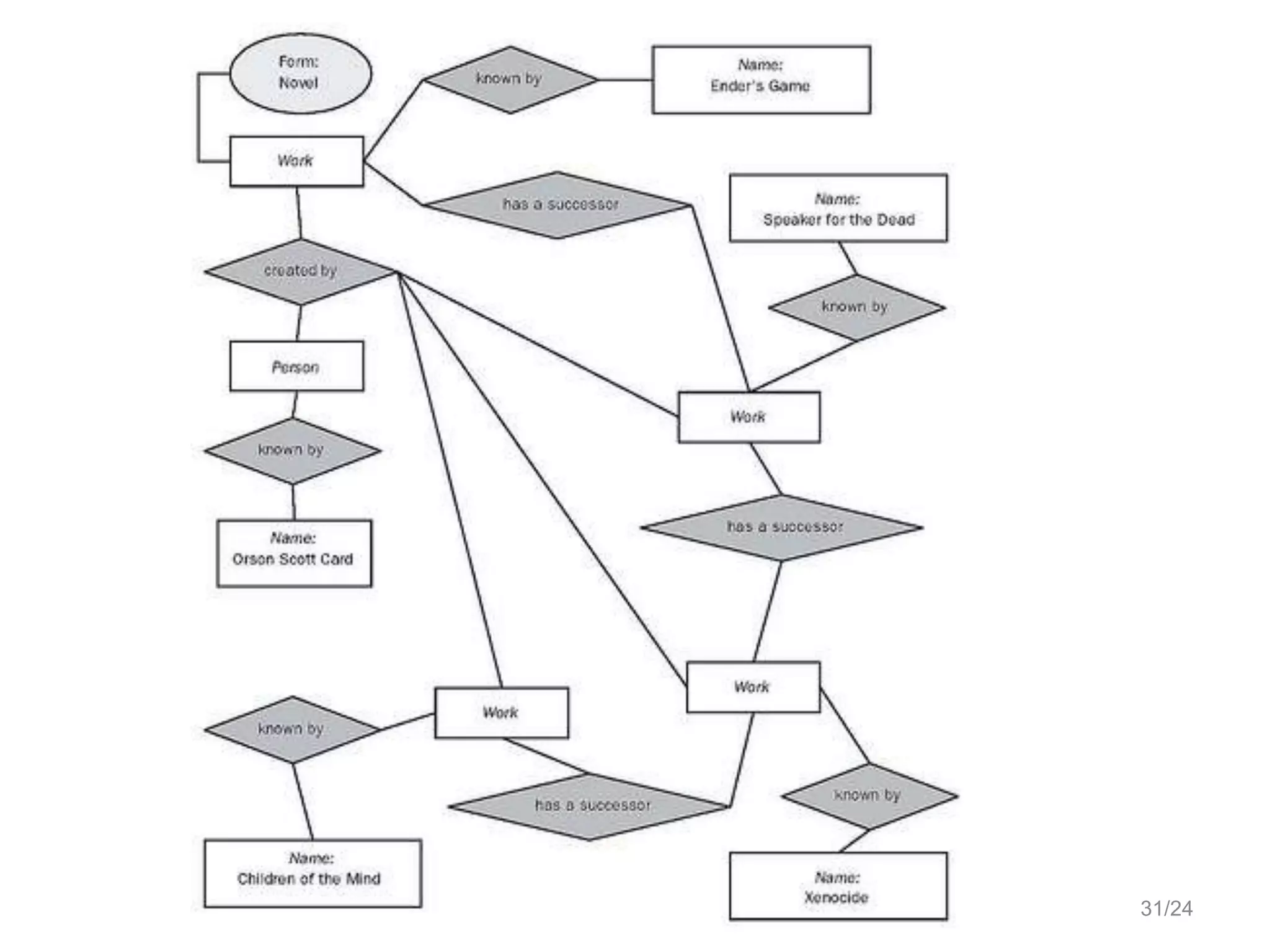
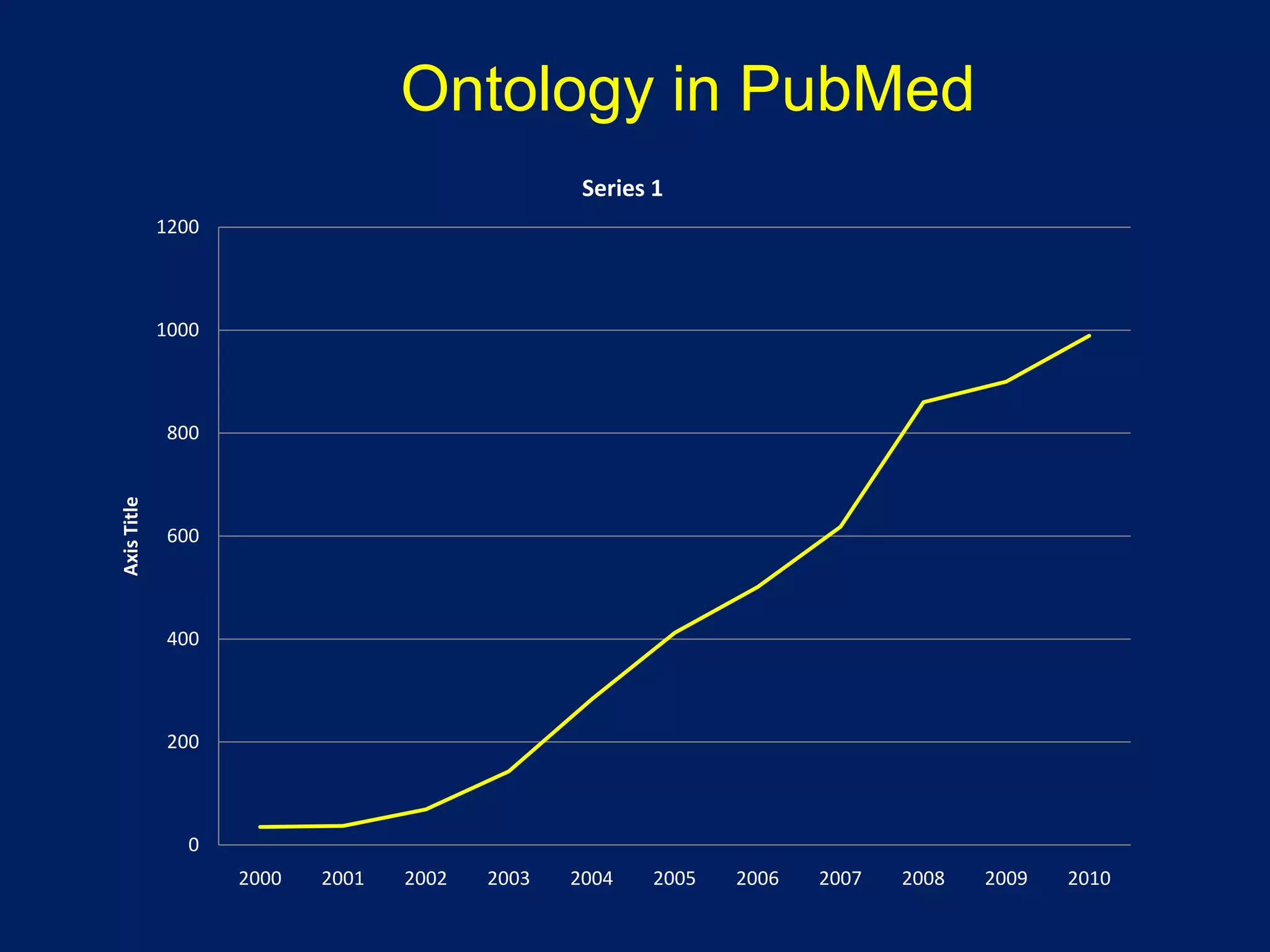
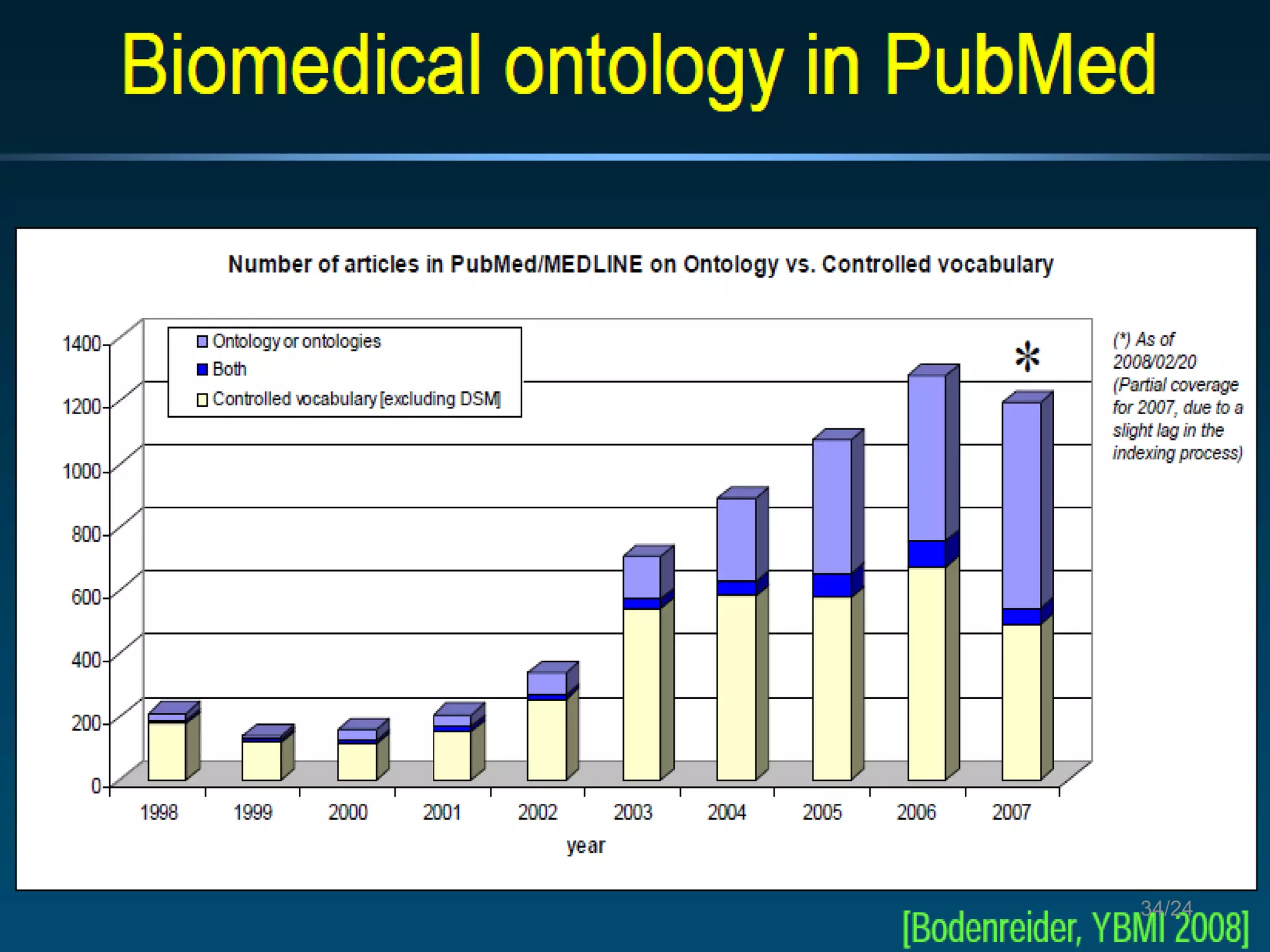
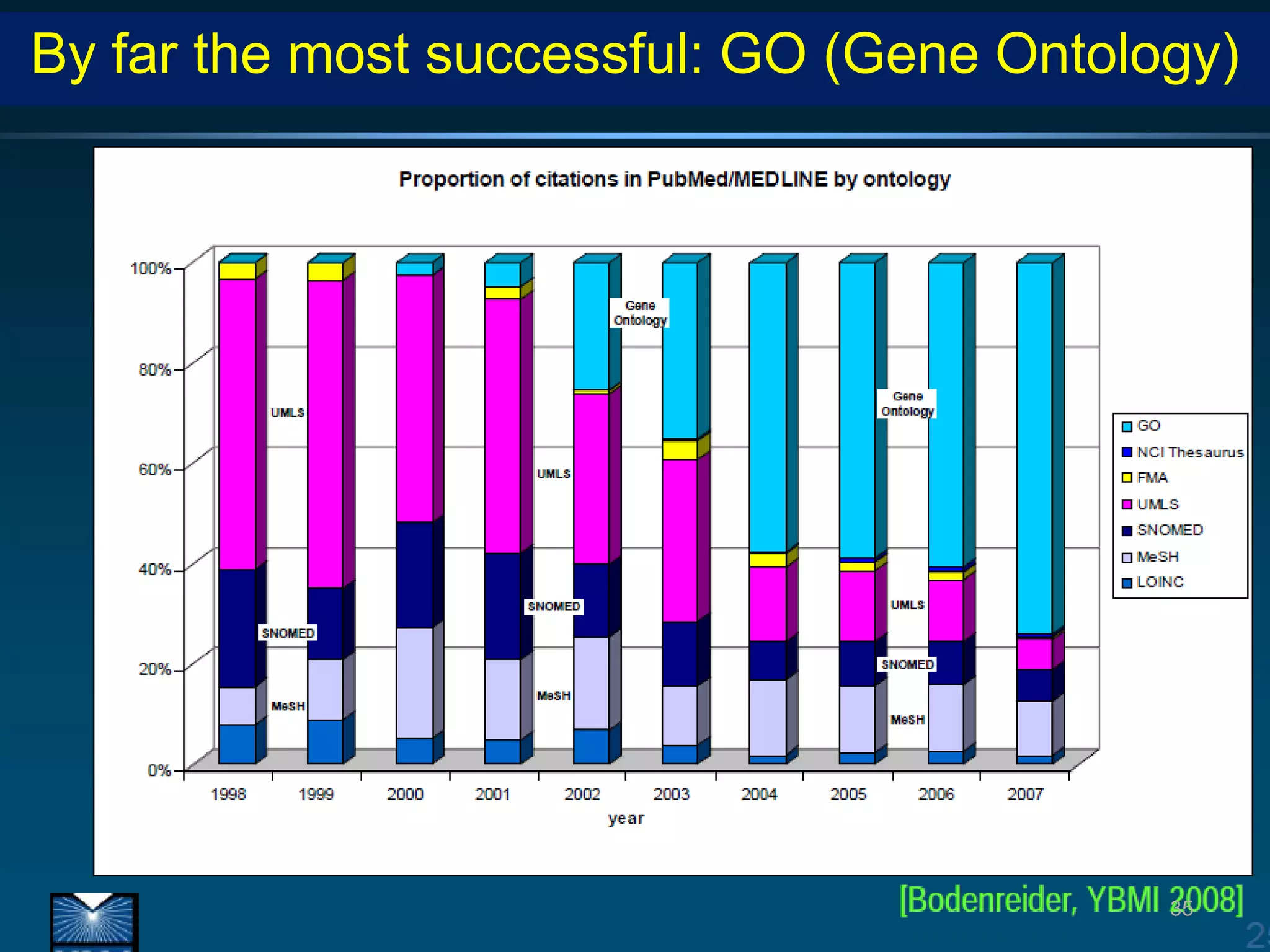
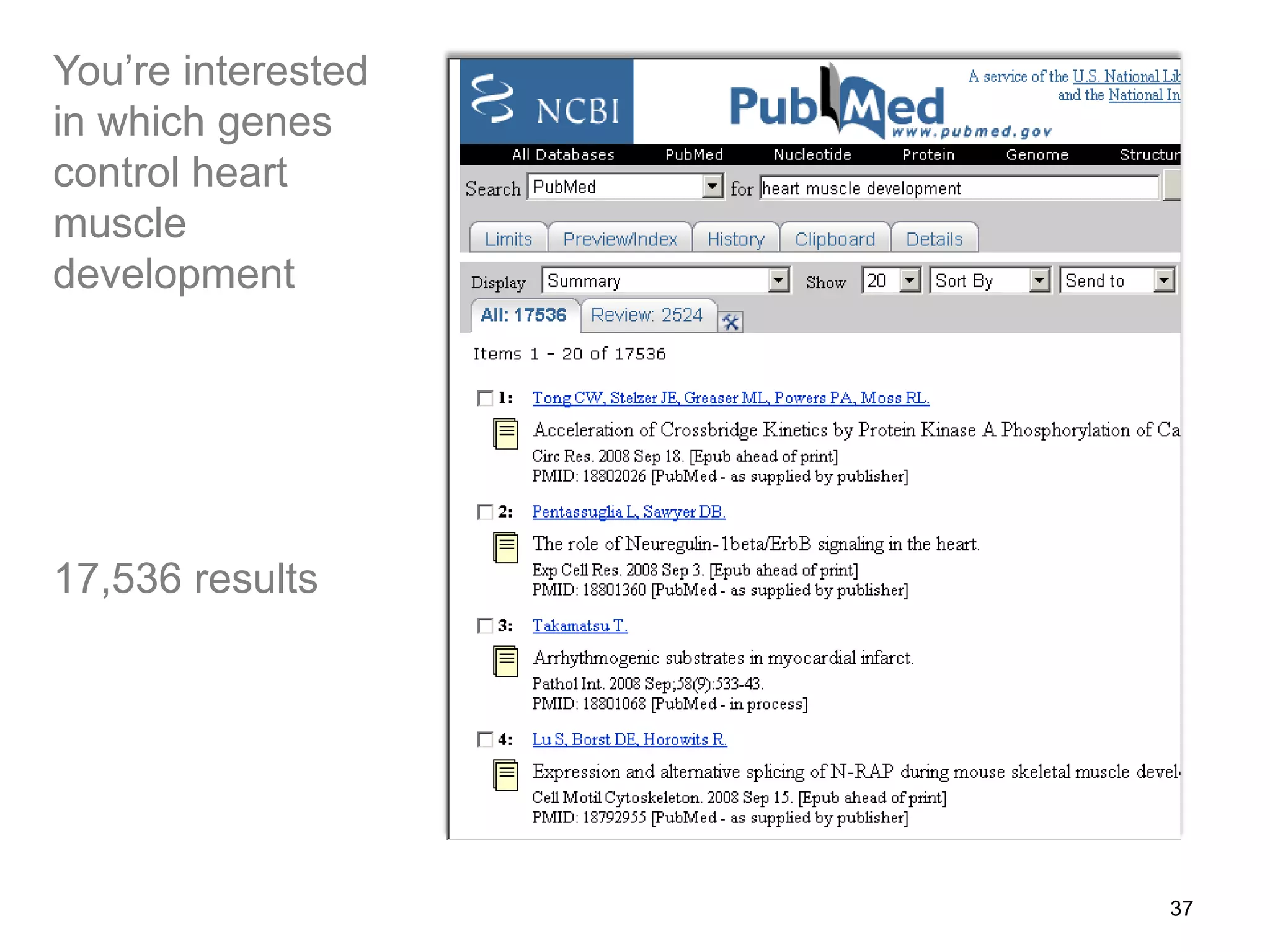
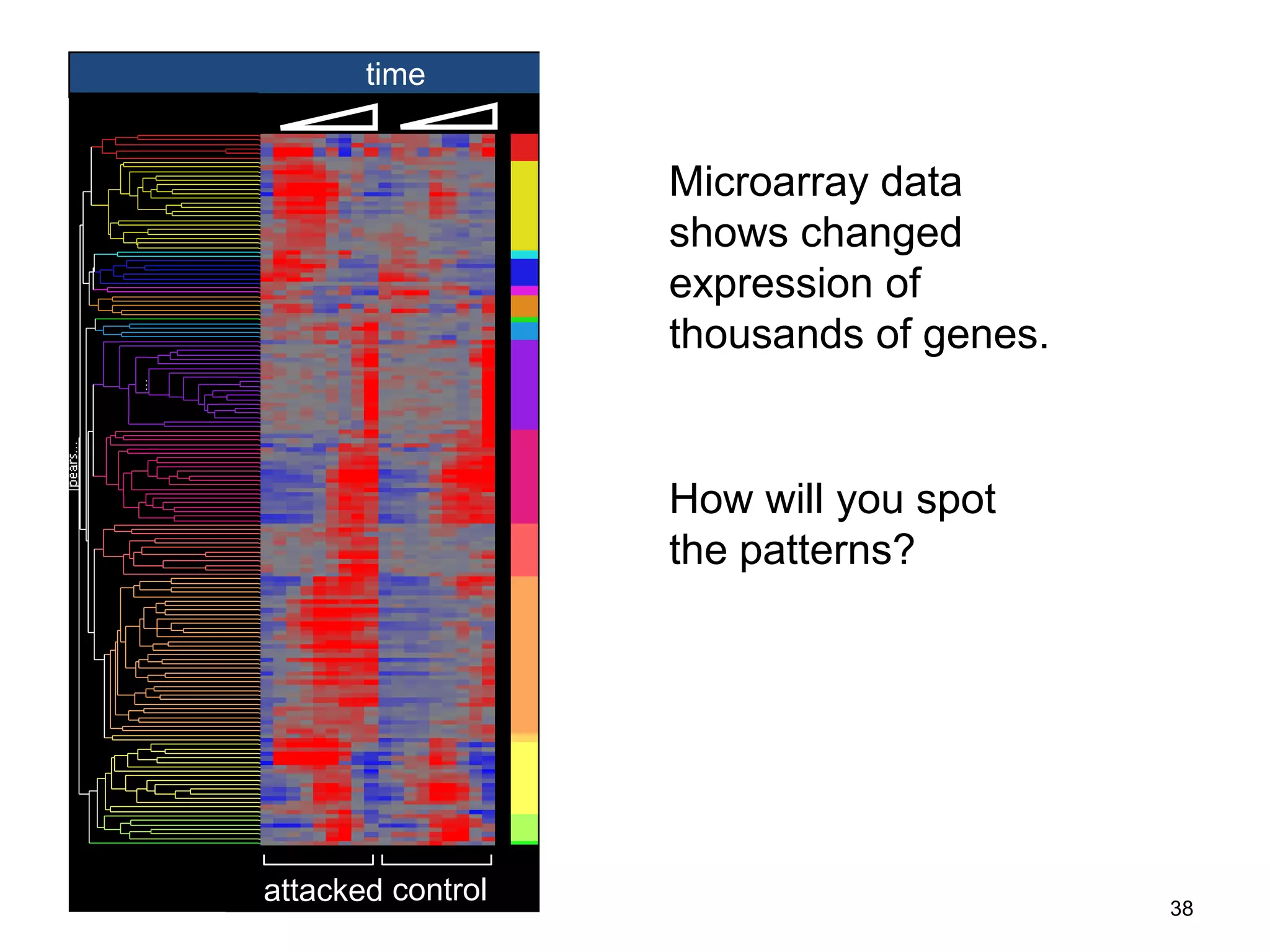
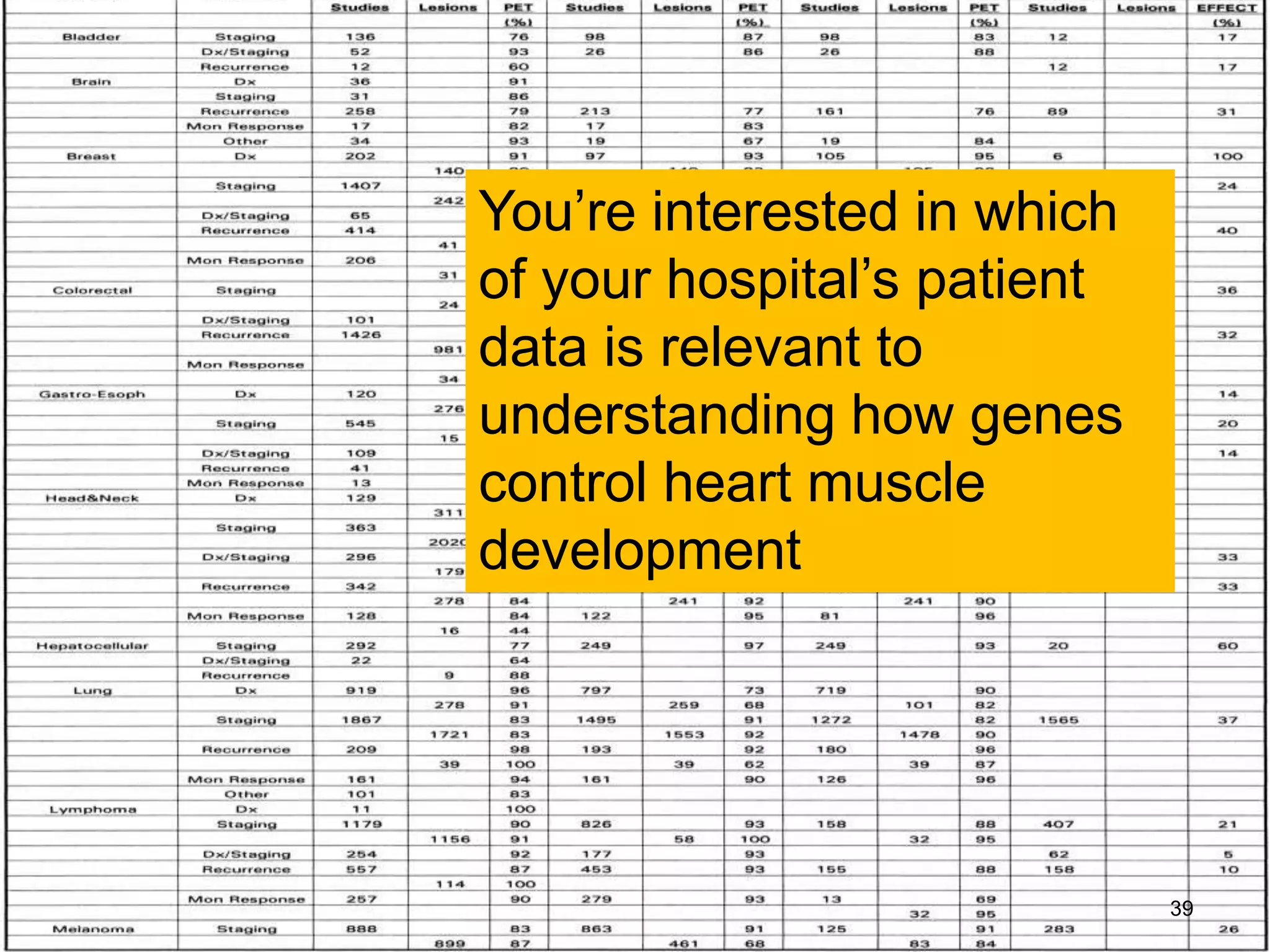
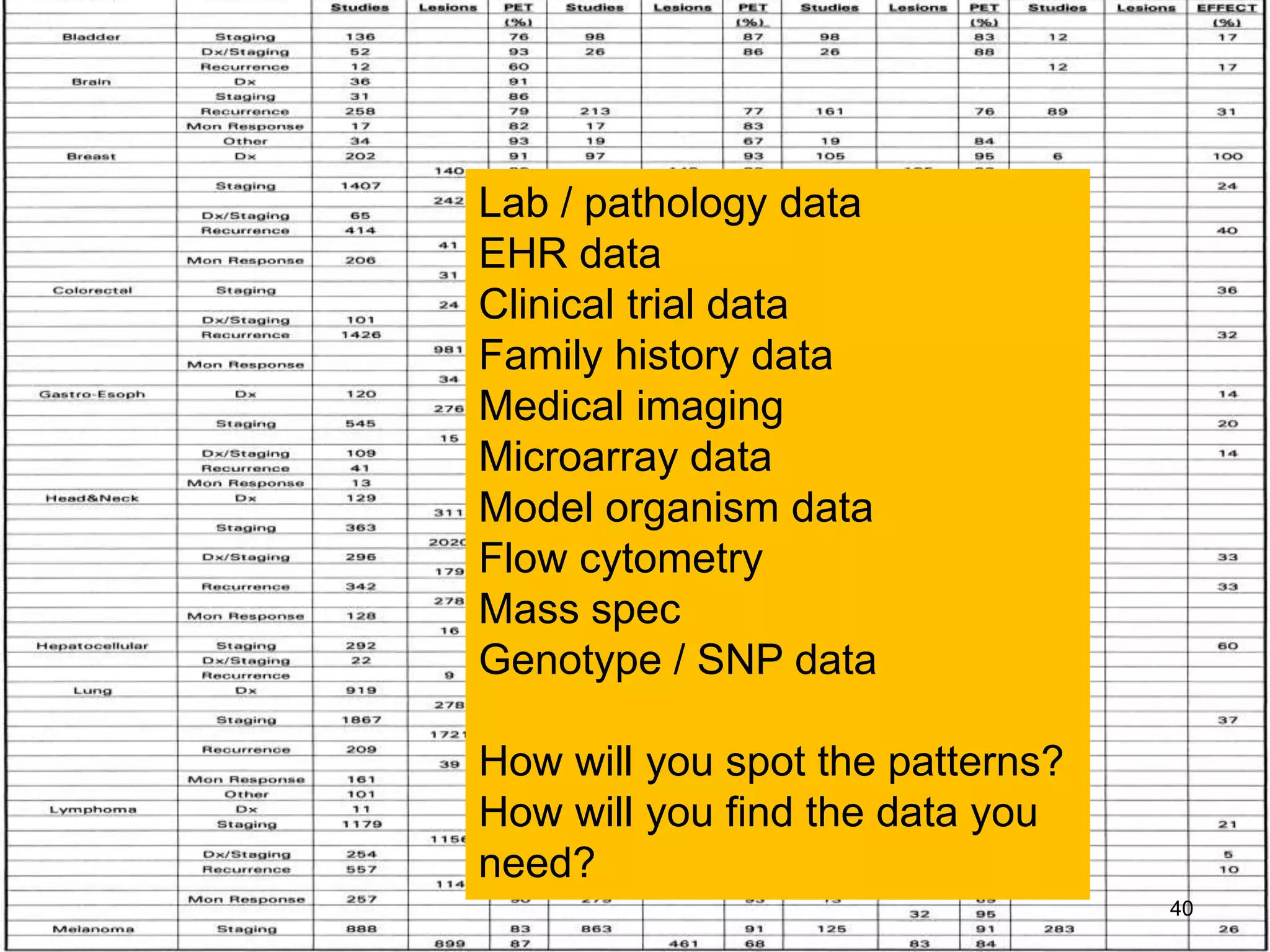
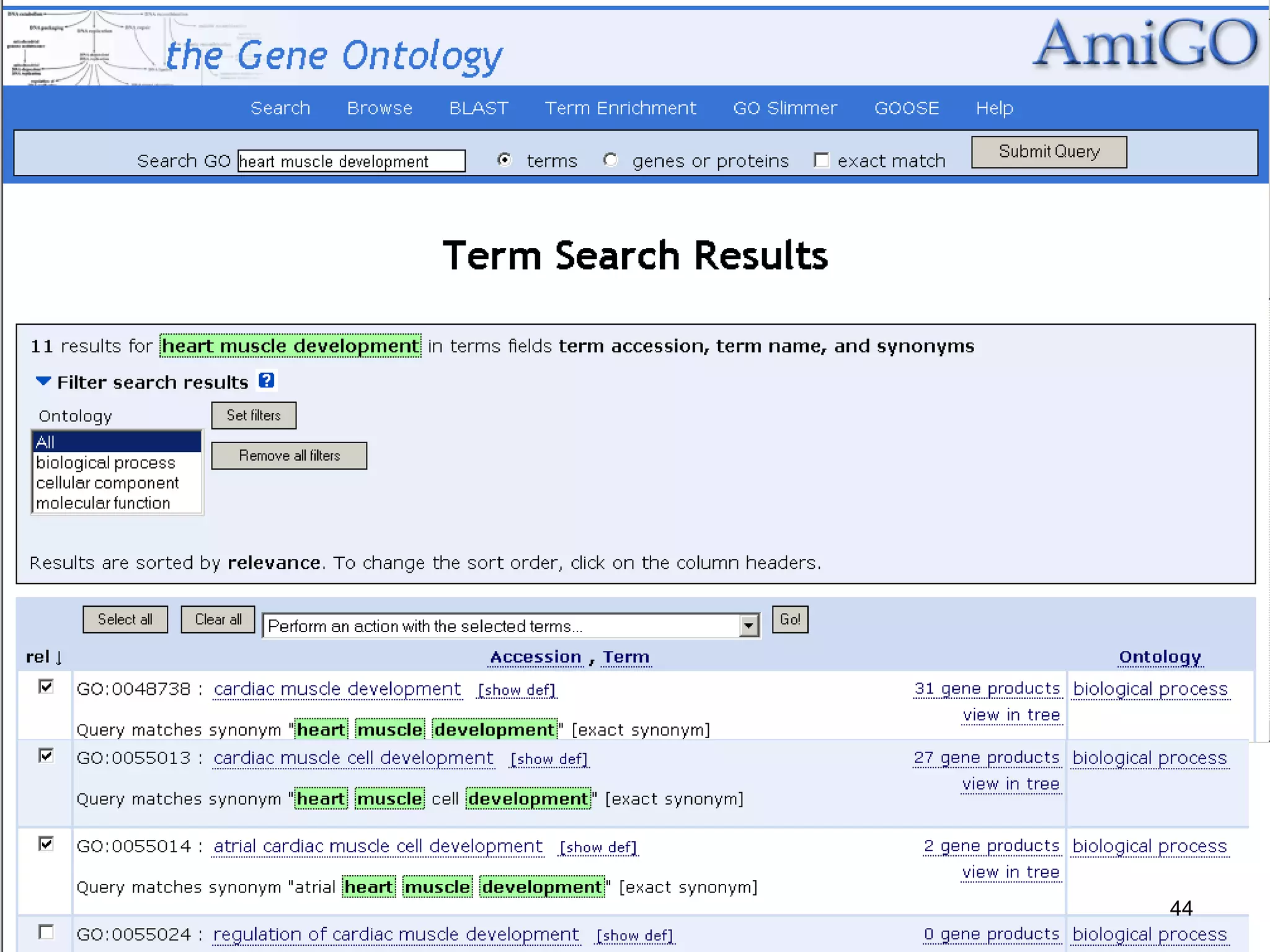
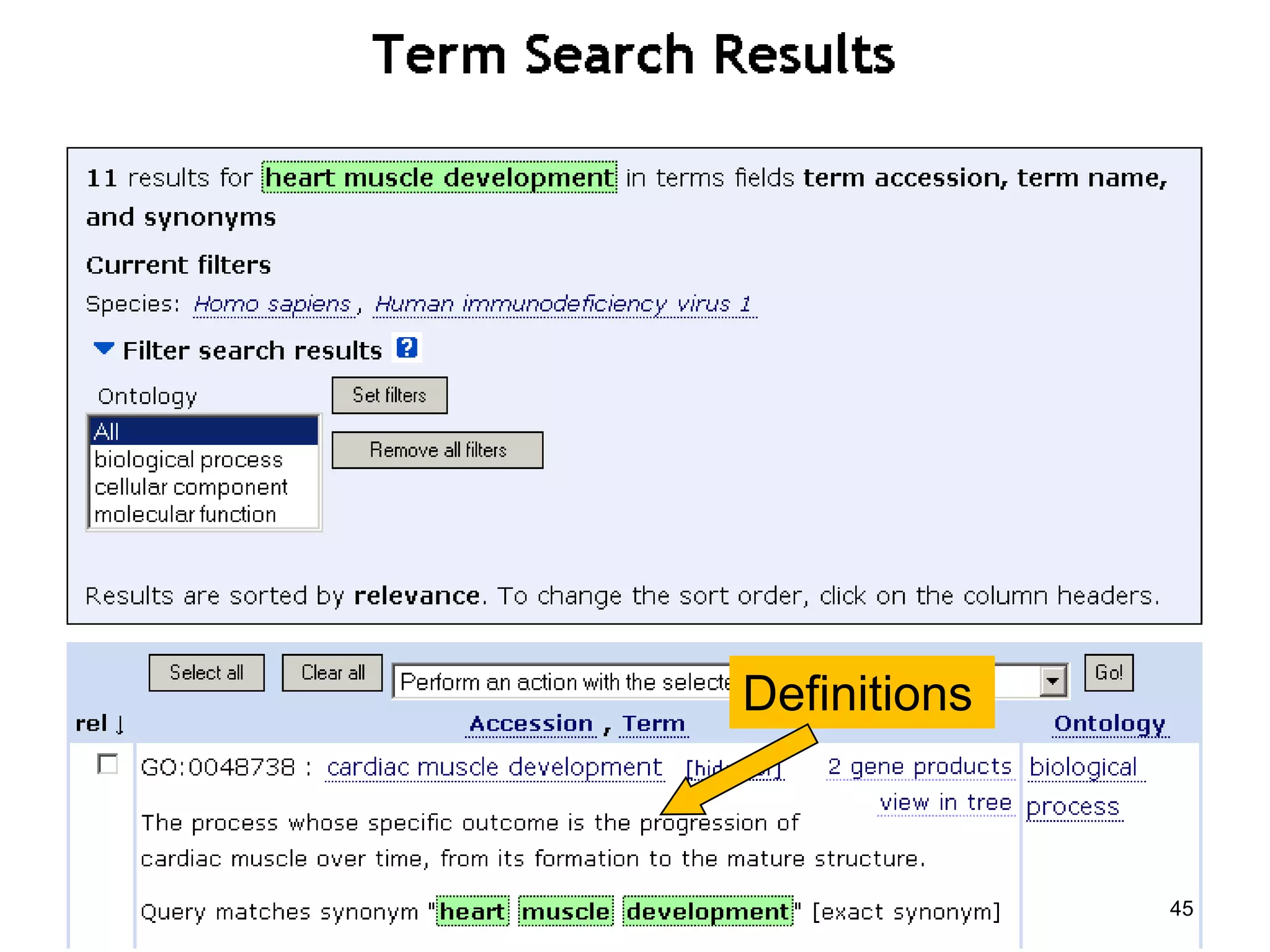
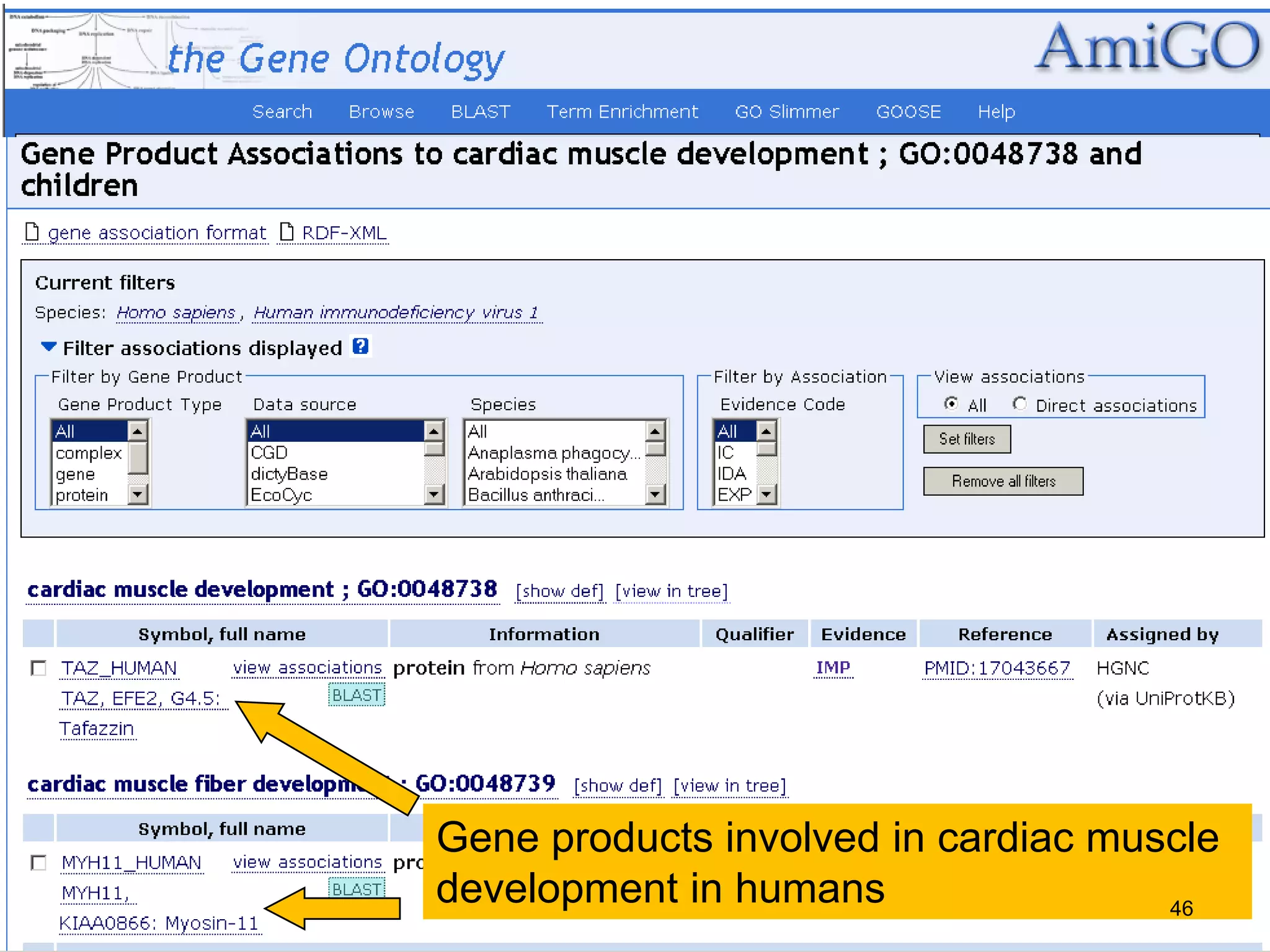
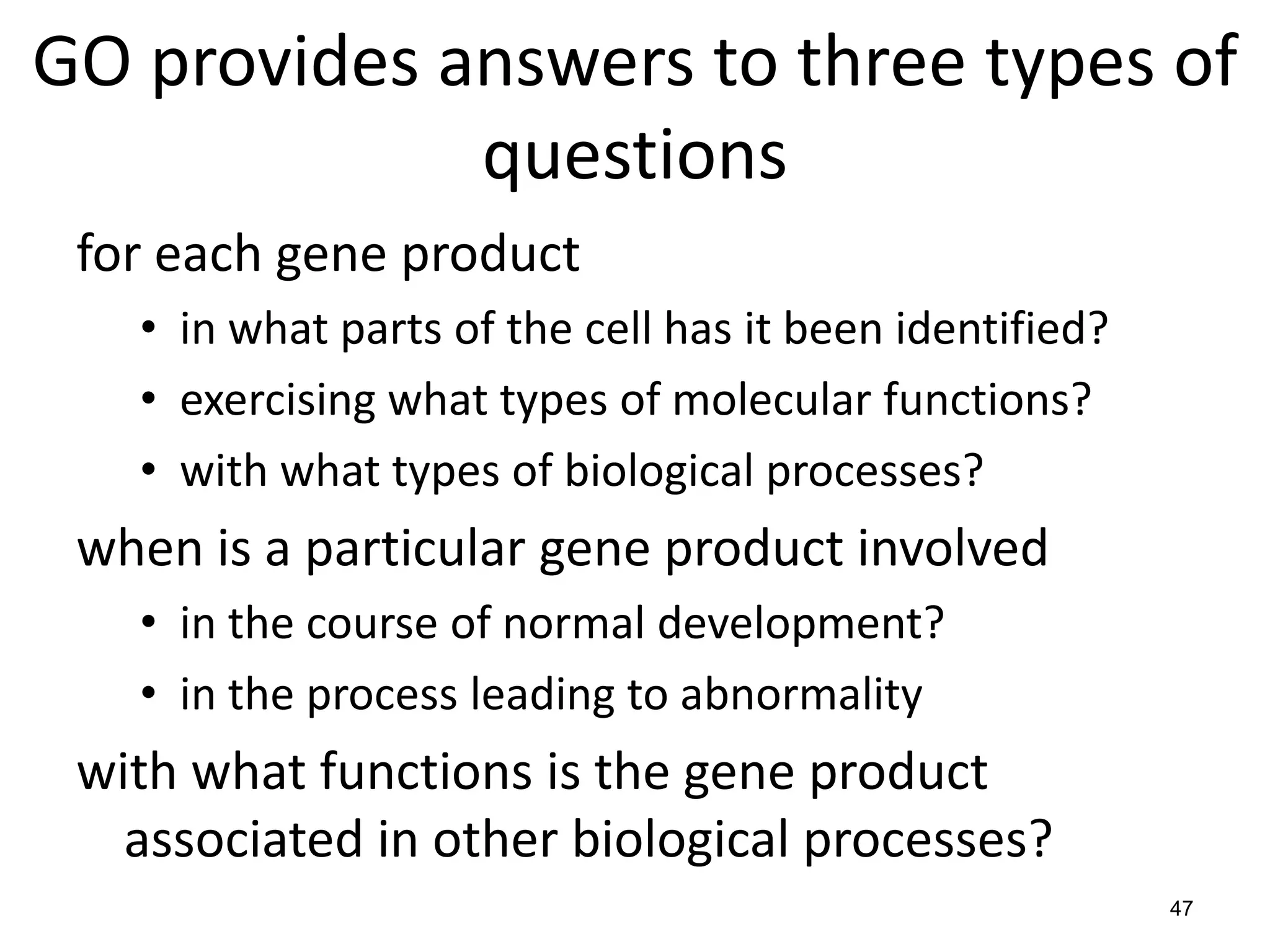
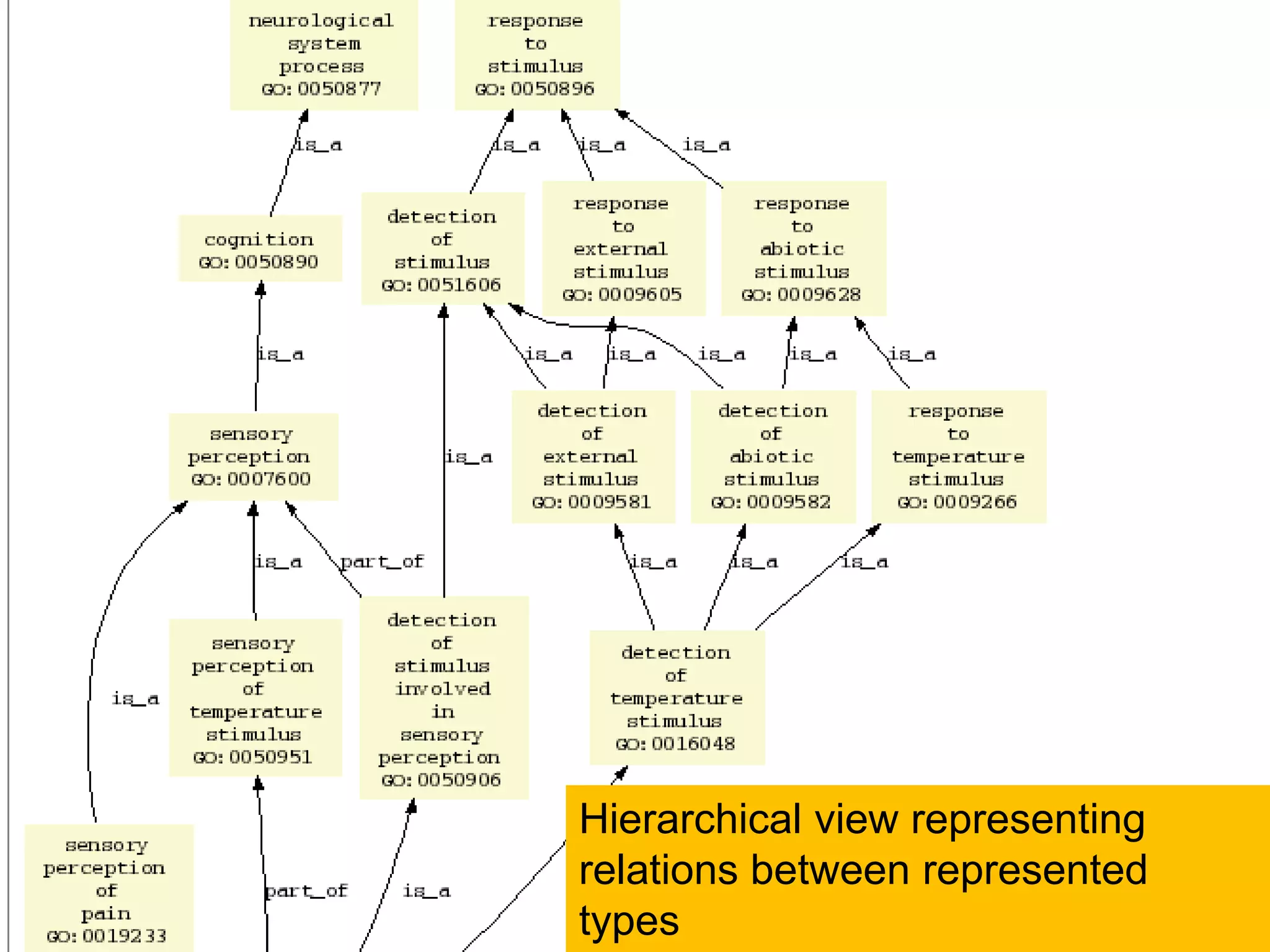
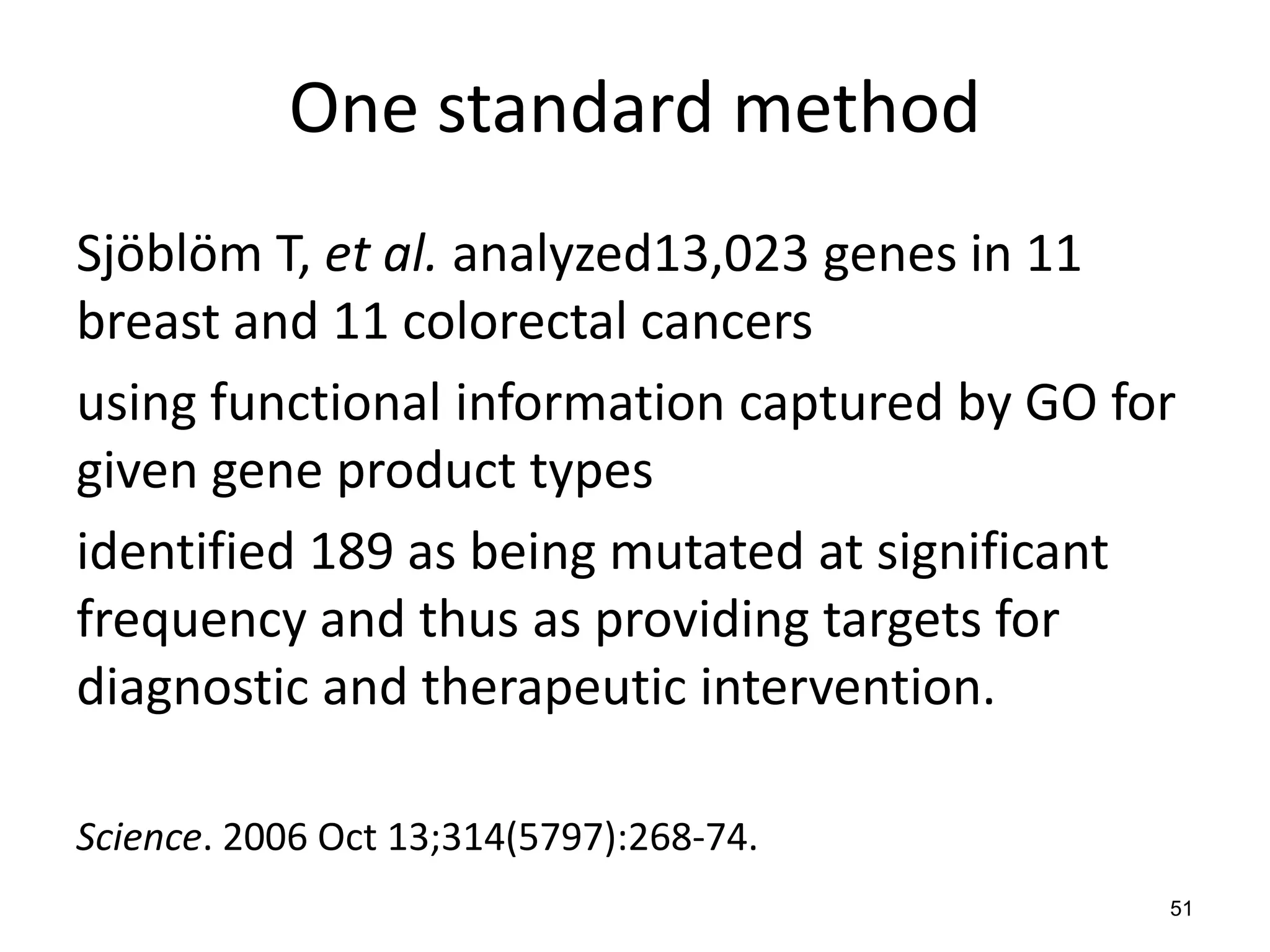
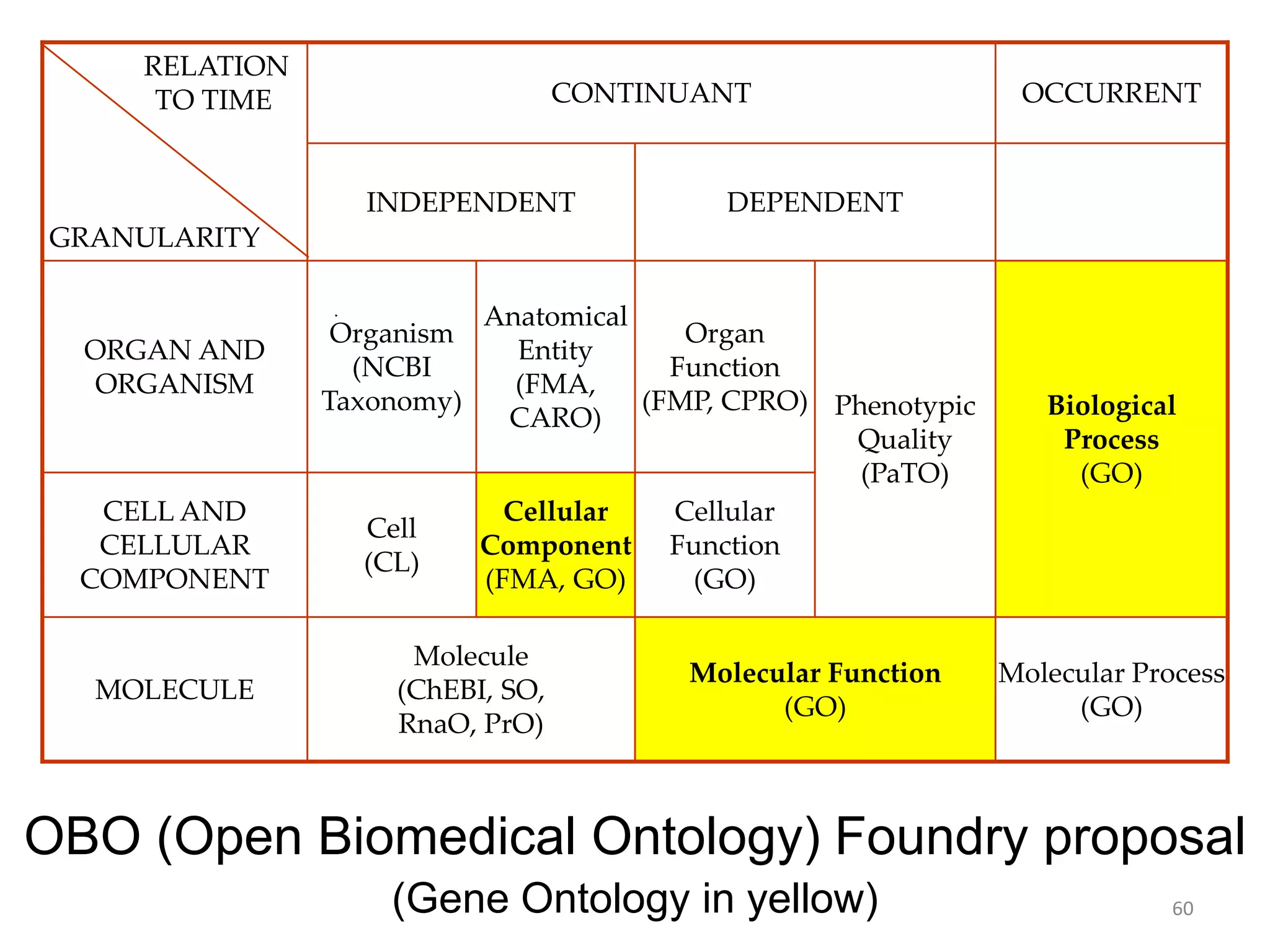
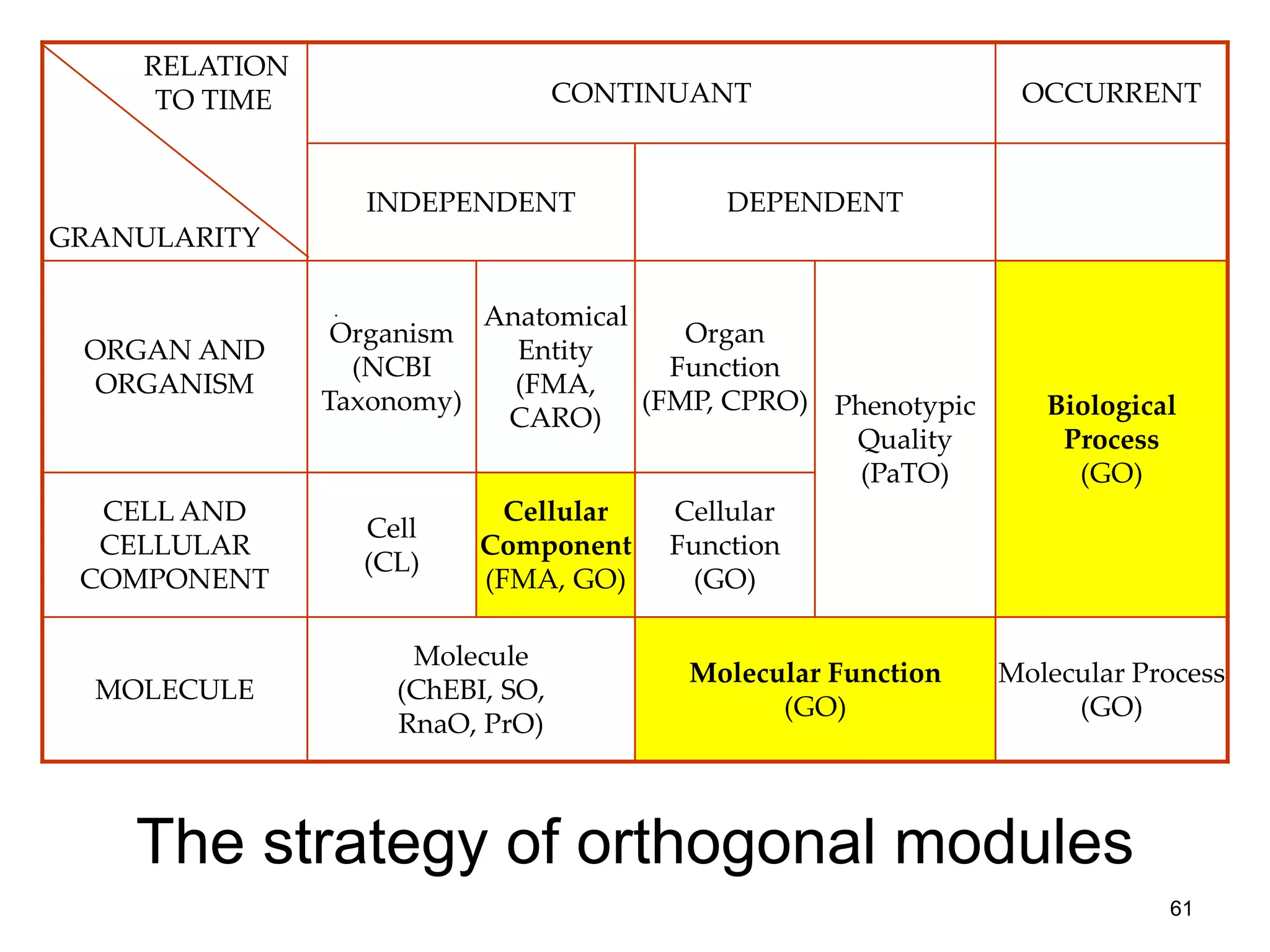
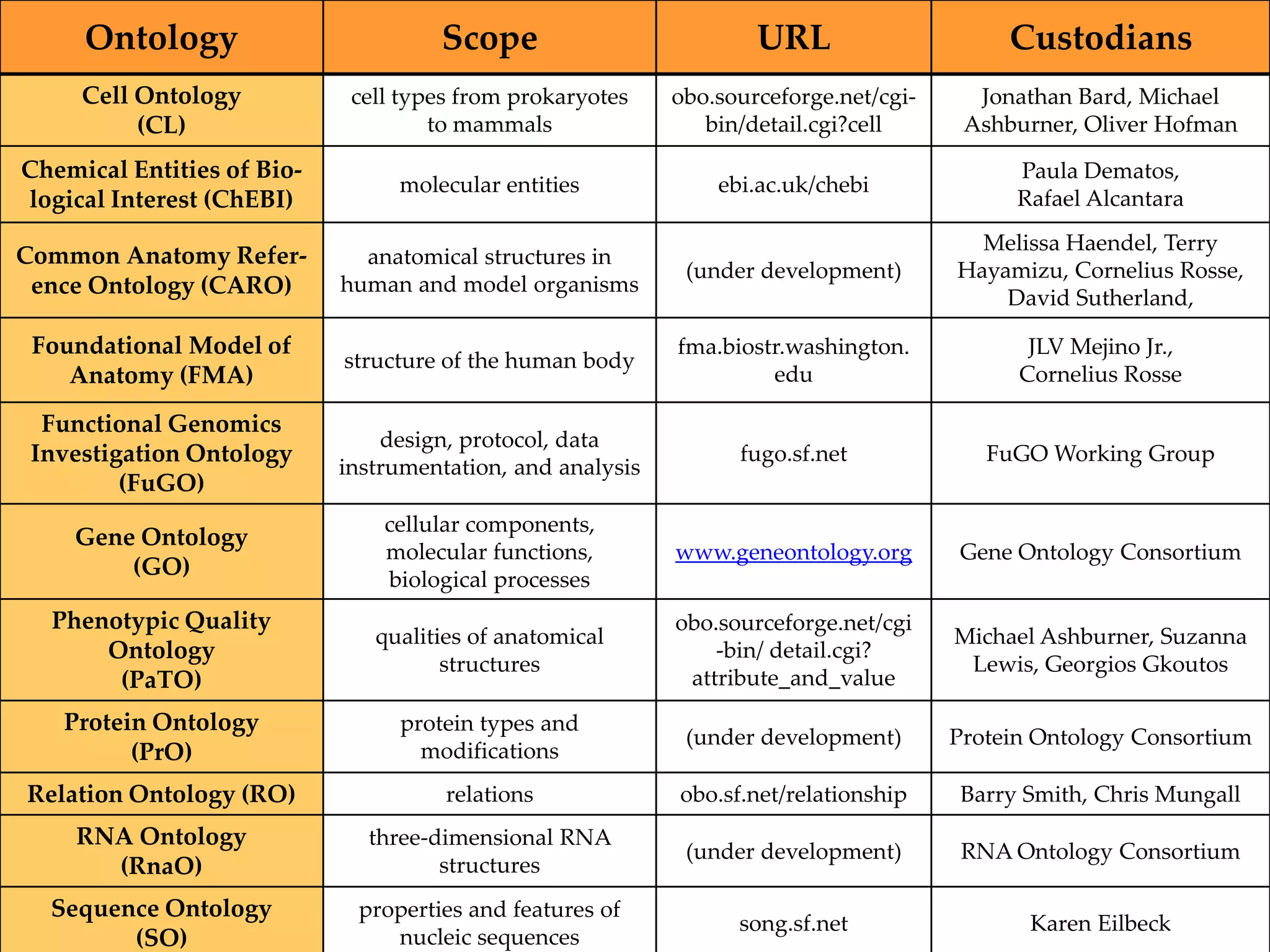
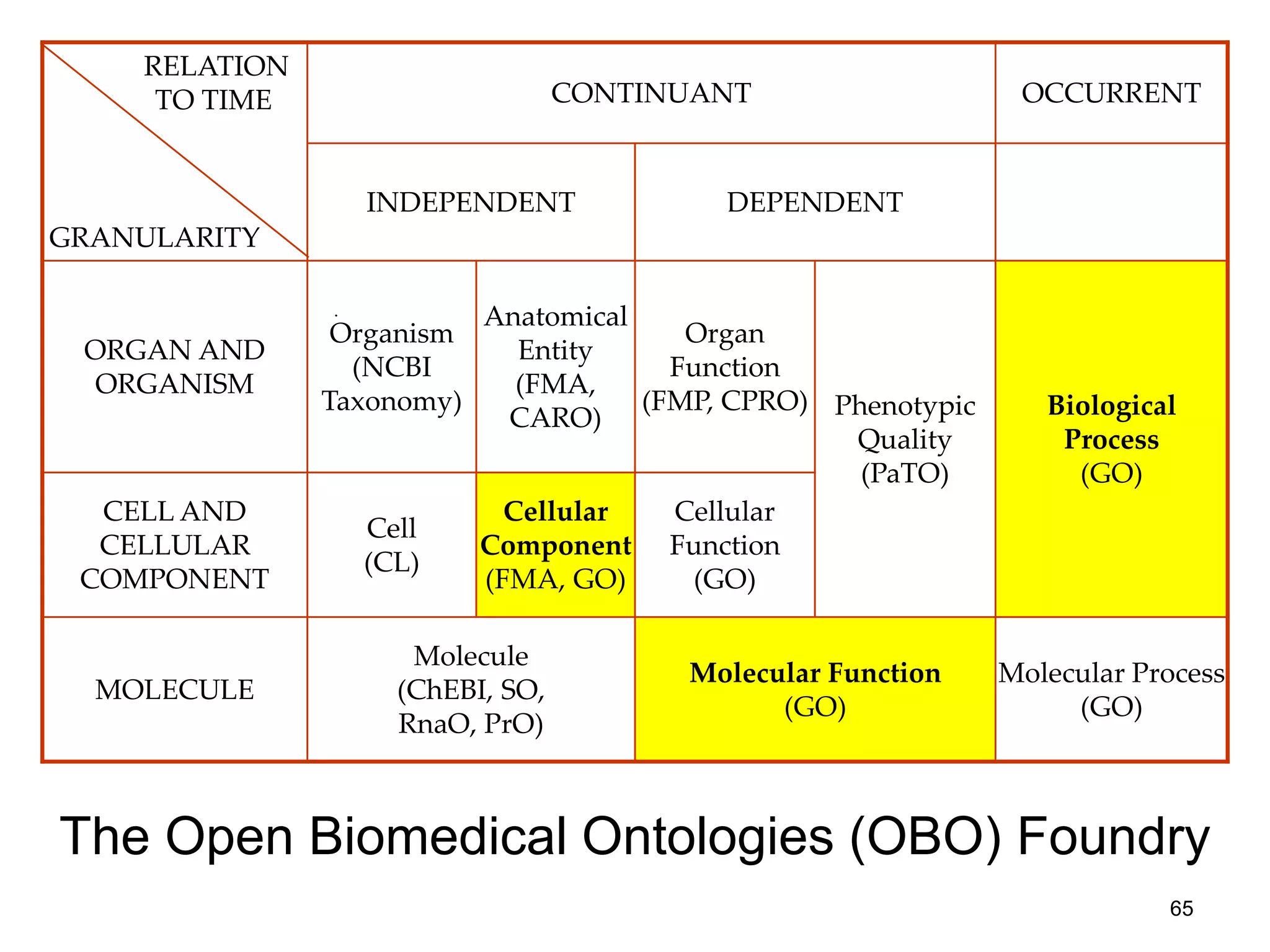
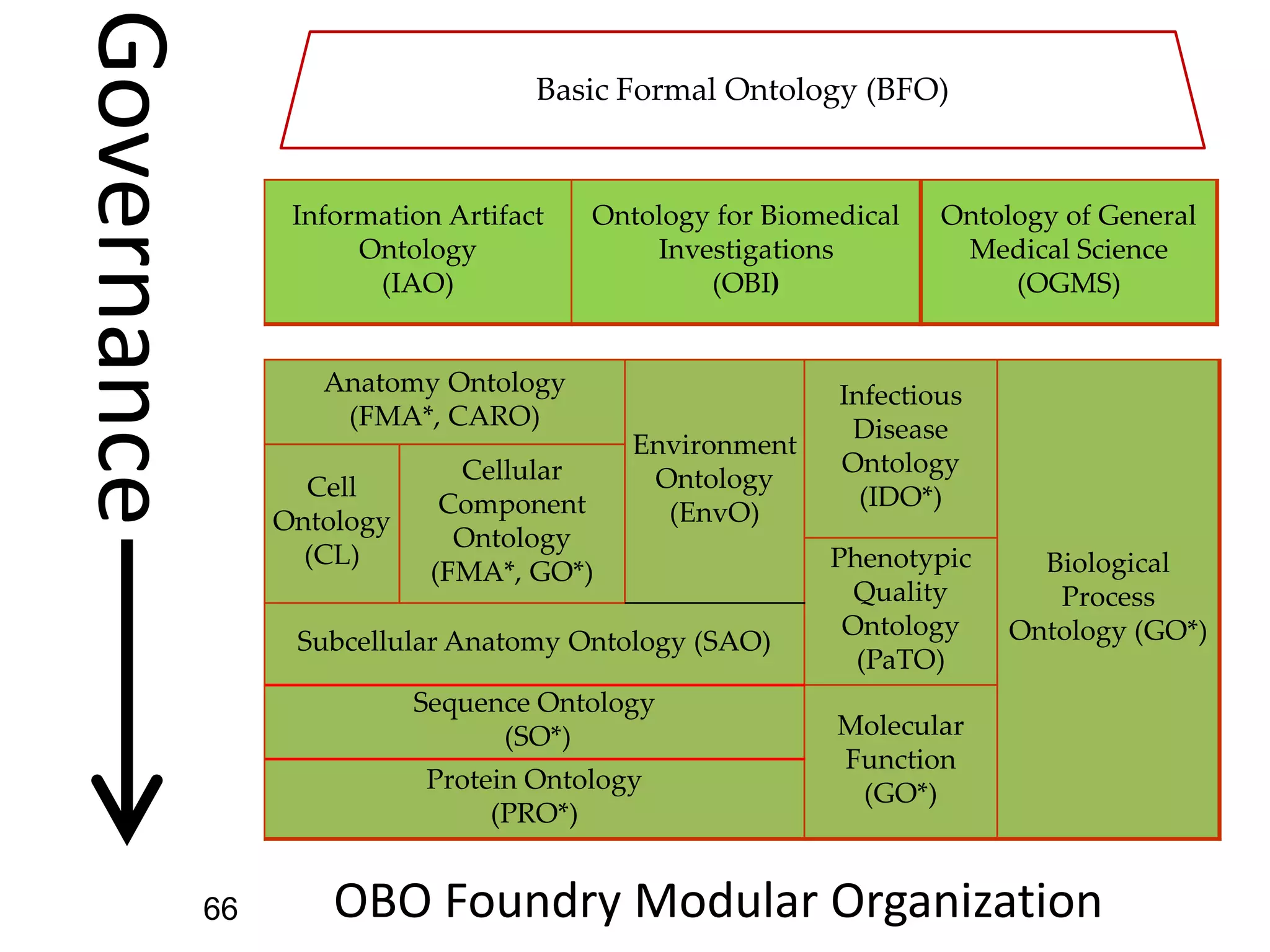
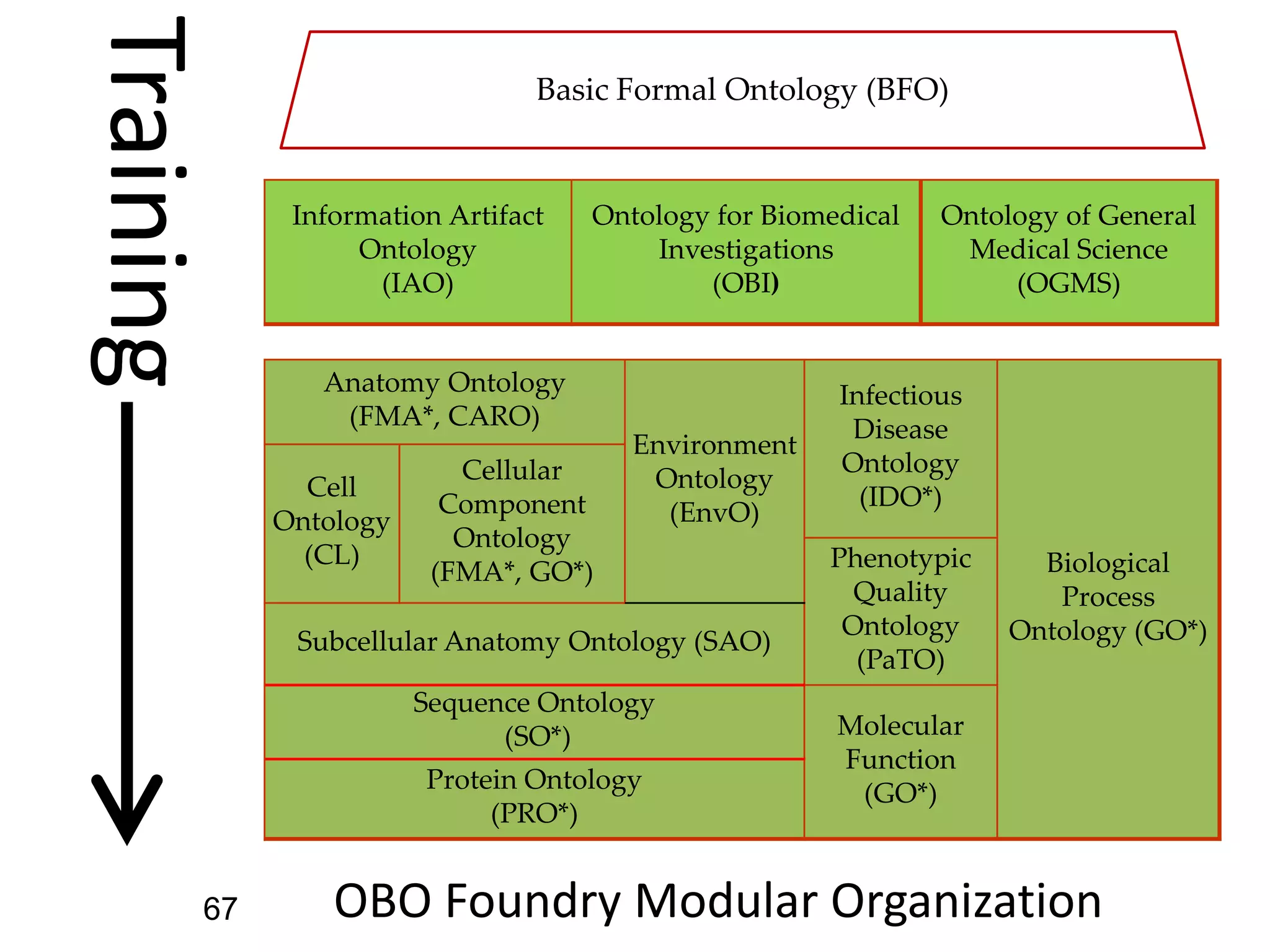
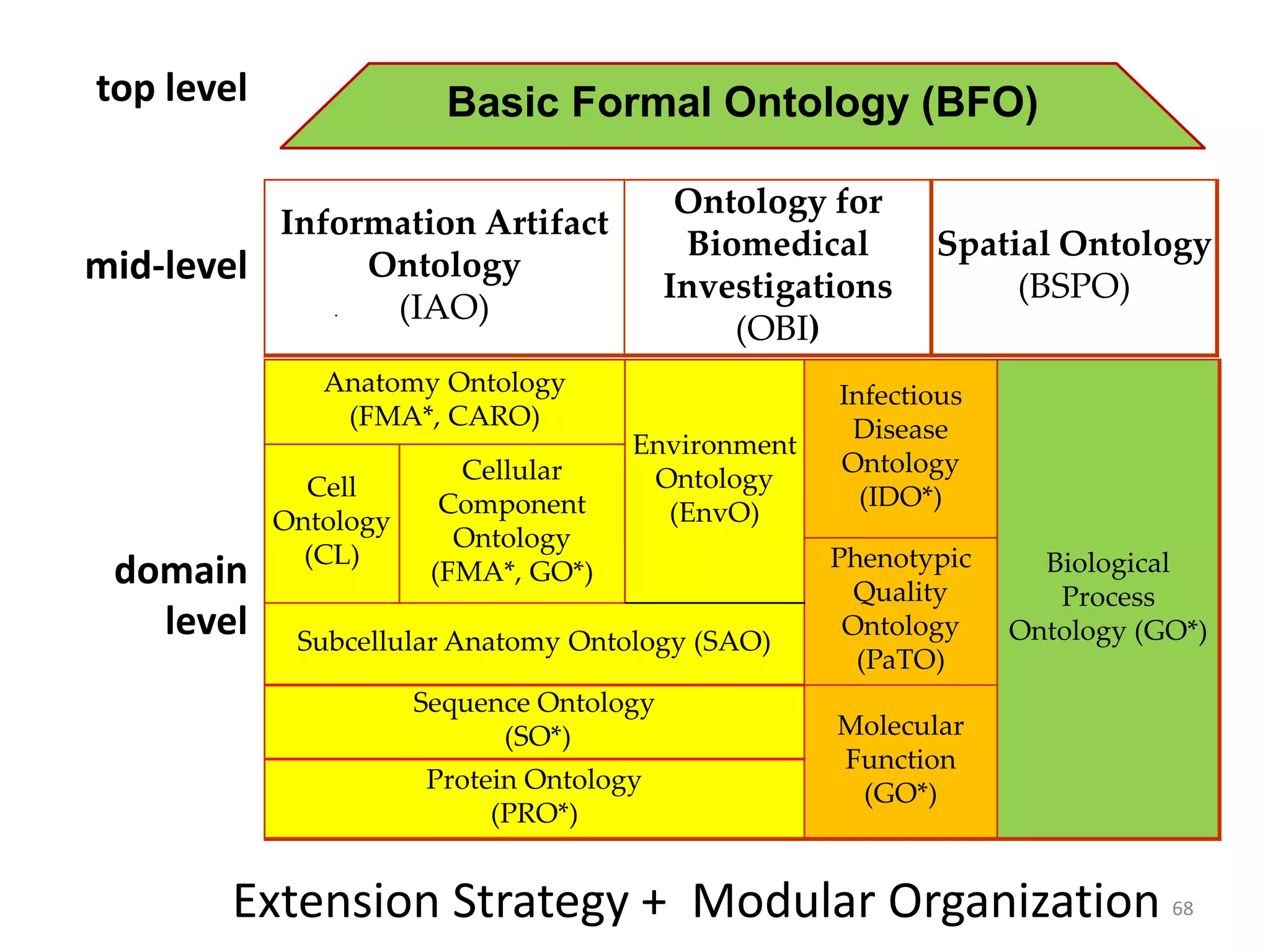
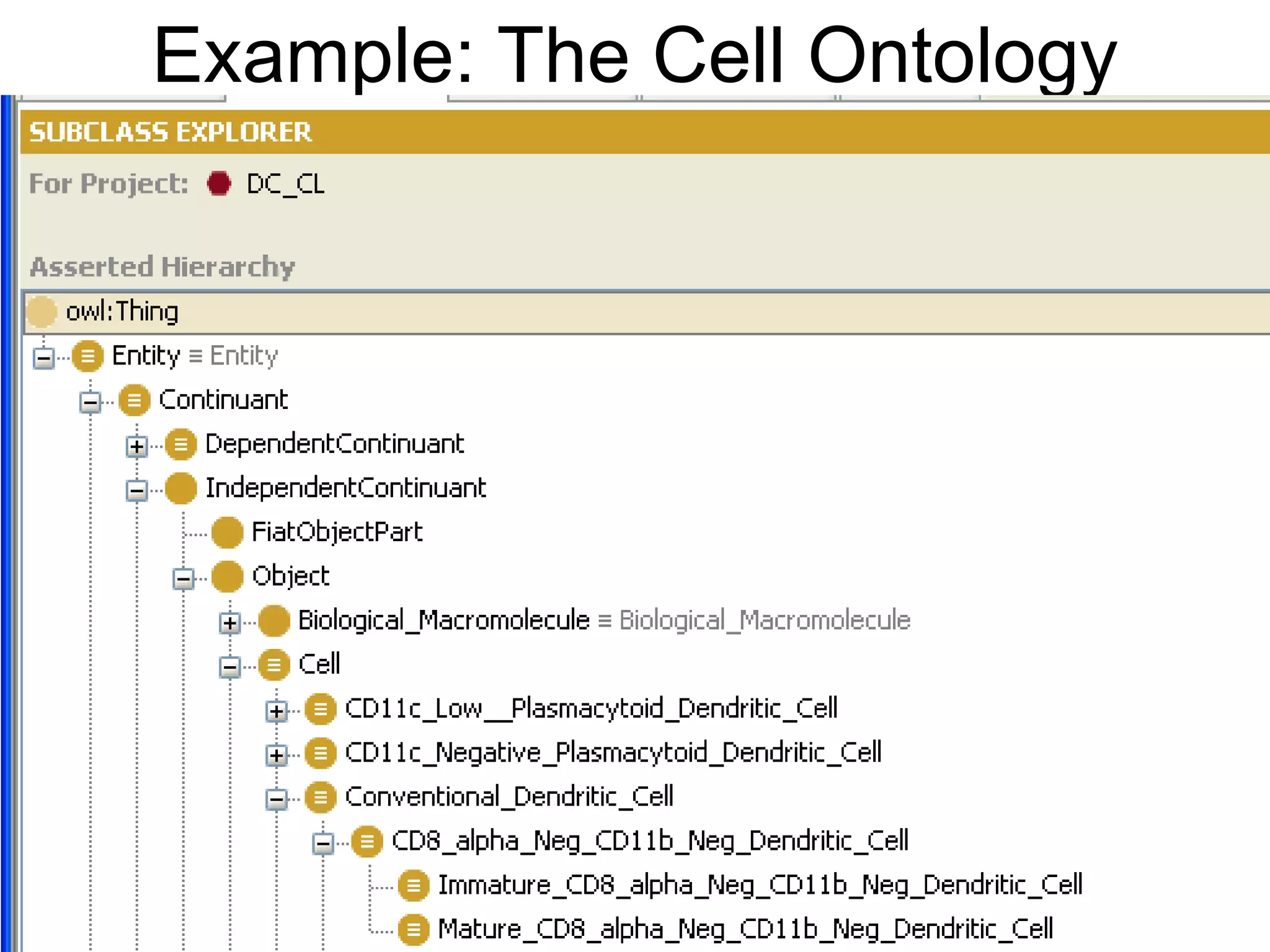
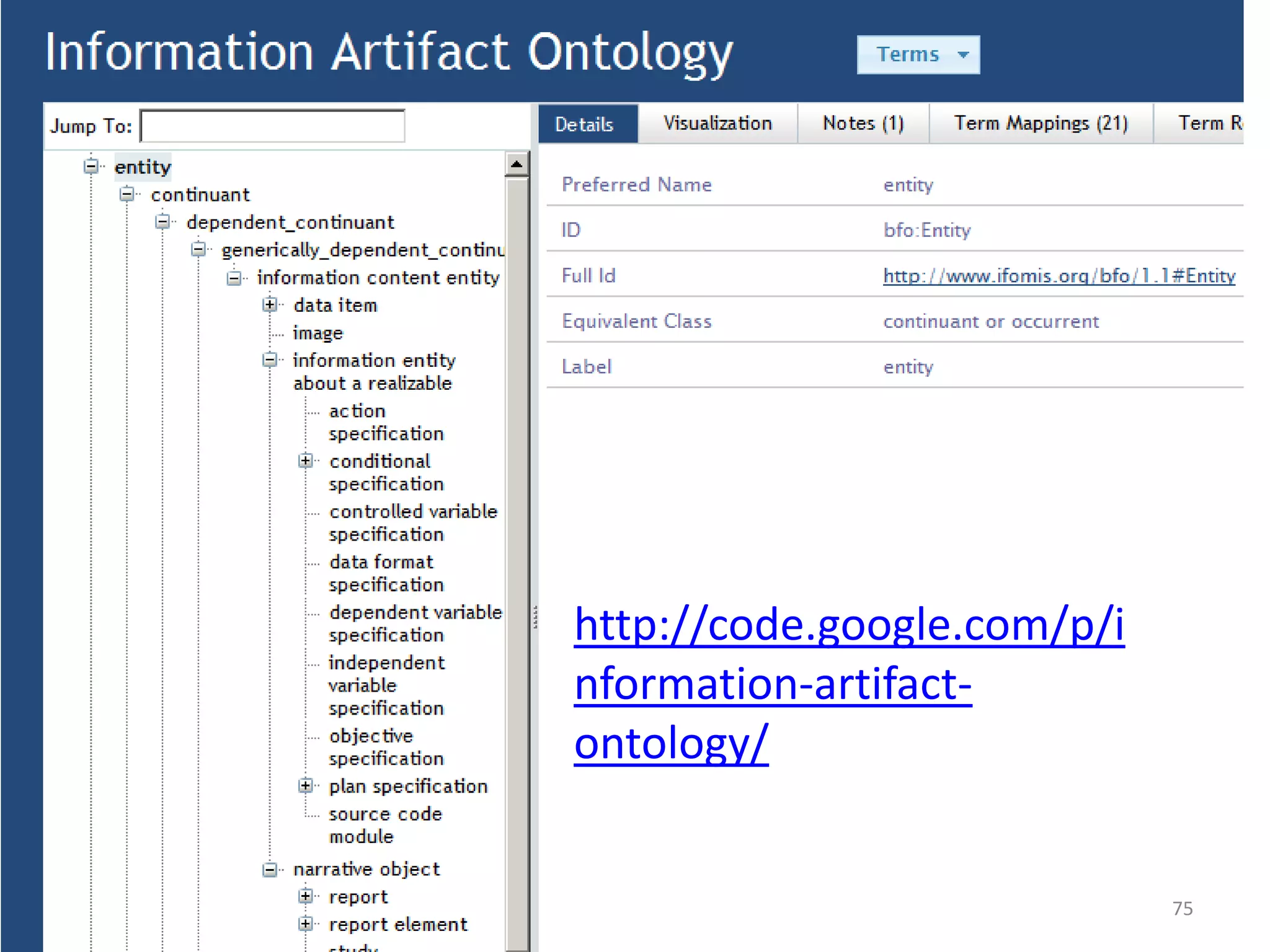
Barry Smith presented on ontologies and what librarians need to know about them. Ontologies provide controlled vocabularies that can be used to tag and annotate data in order to integrate datasets and avoid data silos. The Gene Ontology is highlighted as a successful ontology due to factors such as being developed and maintained by domain experts according to best practices, having over 11 million annotations linking genes to ontology terms, and enabling new types of biological research through analysis and comparison of massive quantities of annotated data. For ontologies to fully realize their potential to remove data silos, they must be prospectively standardized and evolved based on user feedback.