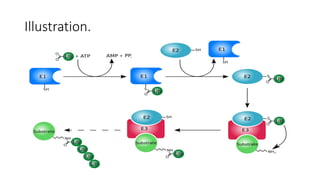
Ubiquitination is a post-translational modification where ubiquitin proteins are attached to substrate proteins. This affects proteins in various ways such as marking them for degradation by the proteasome, altering their cellular location, or promoting or preventing protein interactions. The ubiquitination process involves E1, E2, and E3 enzymes and can result in either mono-ubiquitination with a single ubiquitin attached, or poly-ubiquitination with multiple ubiquitins in a chain. Ubiquitination plays important roles in processes like cell signaling, DNA repair, and the immune response, and dysregulation can contribute to diseases such as neurodegeneration and cancer.