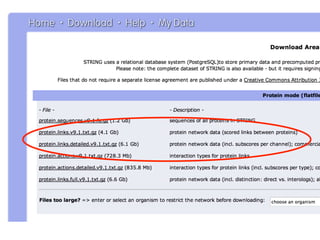
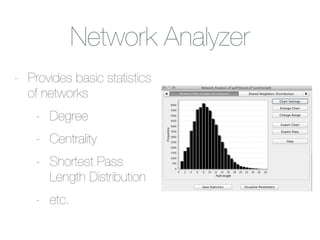
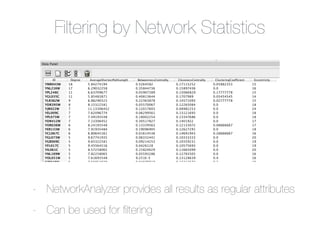
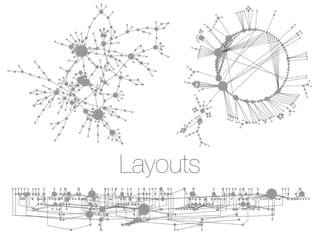
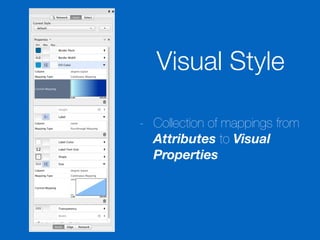
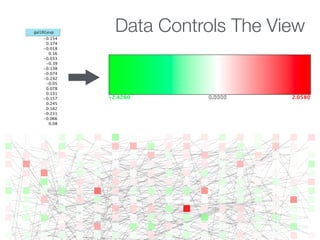
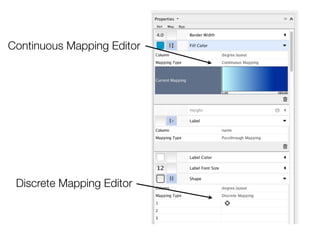
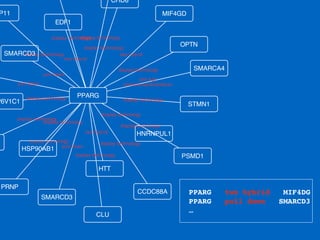
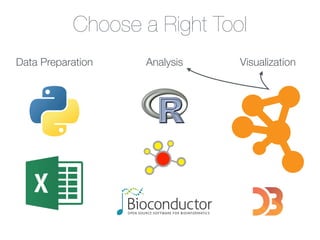
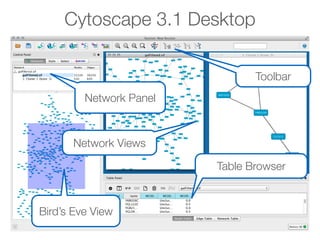
This document outlines a tutorial on biological data analysis and visualization using Cytoscape. The tutorial covers basic concepts like networks and tables in Cytoscape, data import, network analysis features, and visualization techniques. It discusses loading sample network data, calculating network statistics, filtering networks, basic search functionality, and applying visual styles. The tutorial is intended to provide a practical introduction to Cytoscape's core features through examples and demos.







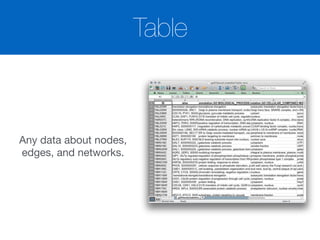







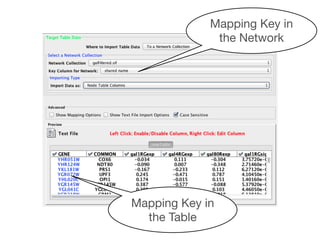
![Table Browser
Local Column
Table Tabs
List Data
(Values in [ ])
Shared Column](https://image.slidesharecdn.com/bioinformaticssummittutorial1-140411191733-phpapp01/85/Cytoscape-Tutorial-Session-1-at-UT-KBRIN-Bioinformatics-Summit-2014-4-11-2014-24-320.jpg)