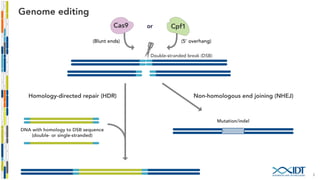
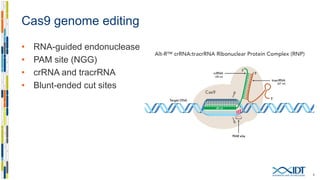
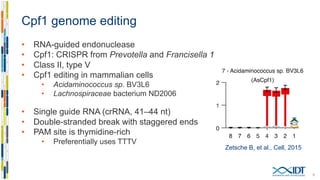
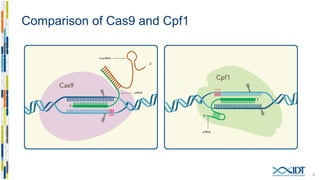
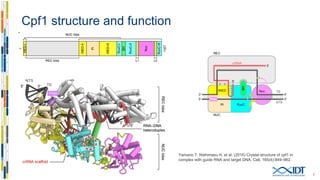
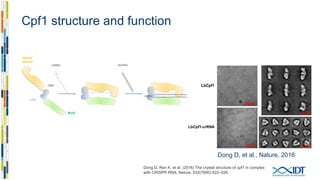
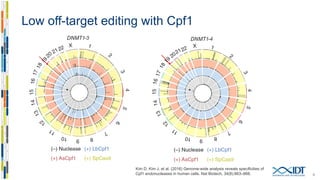
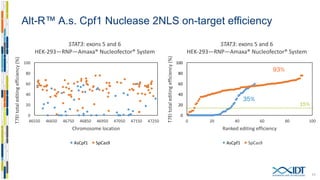
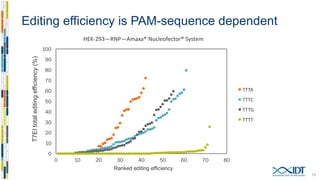
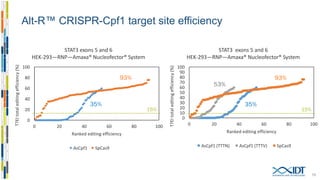
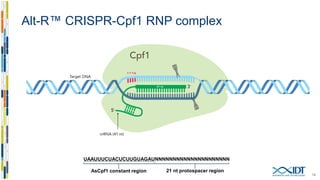
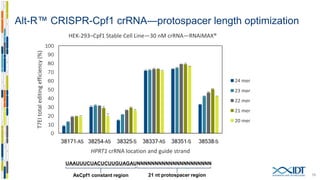
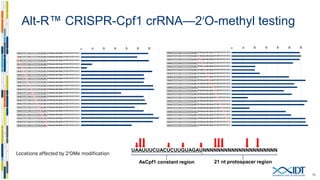
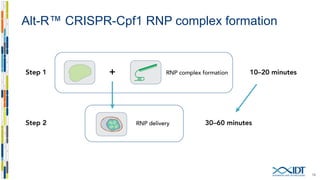
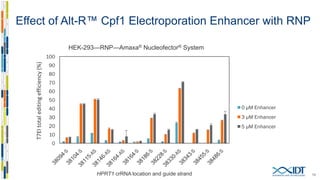
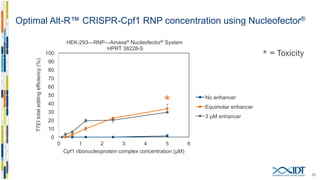
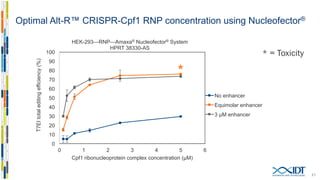
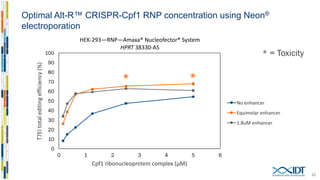
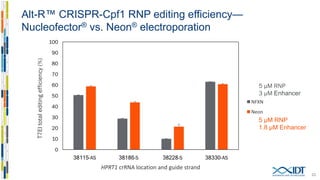
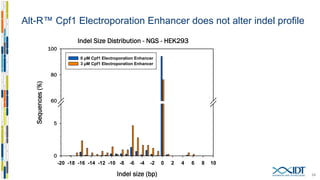
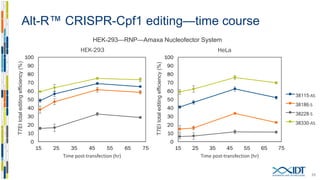
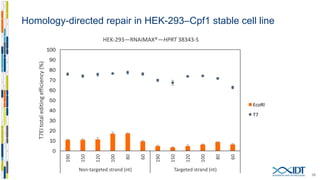
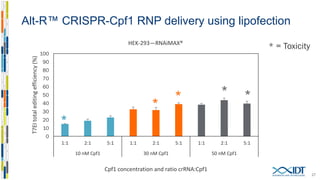
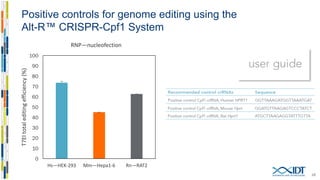
The document discusses the optimization and applications of the alt-RTM CRISPR-Cpf1 genome editing system, covering various aspects such as crRNA optimization, delivery methods as RNP complexes, and homology-directed repair in mammalian cells. It compares the efficiency and specificity of Cpf1 to Cas9 and highlights the advantages of Cpf1, including lower off-target effects and dependency on PAM site sequences. The document also provides data on various experiments conducted to evaluate editing efficiency and methods for enhancing delivery through electroporation.