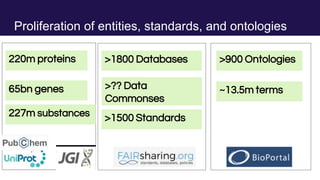
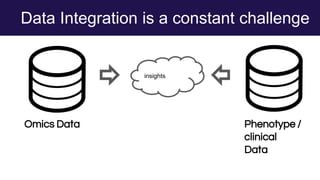
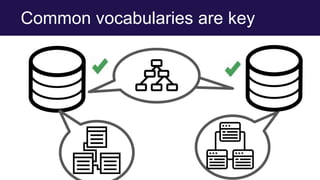
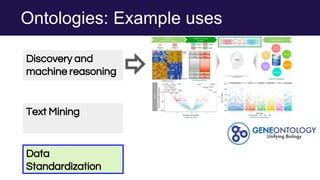
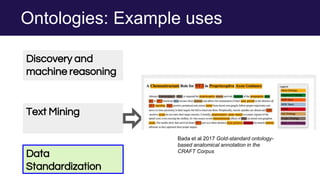
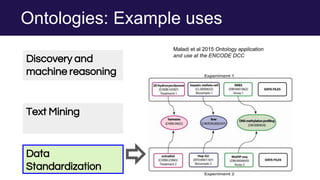
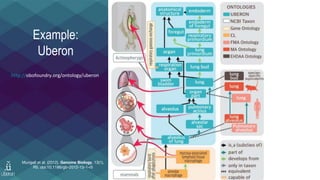
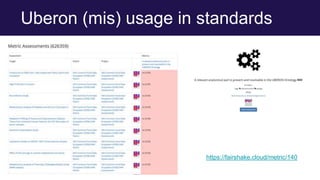
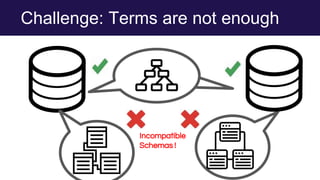
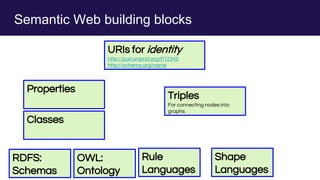
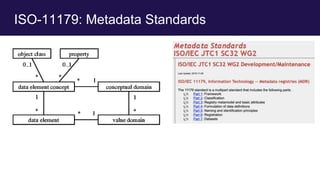
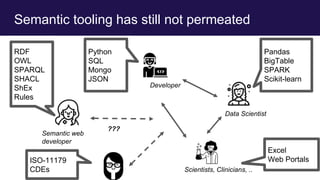
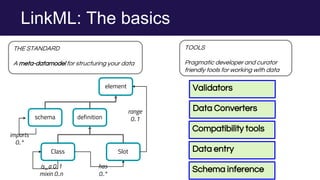
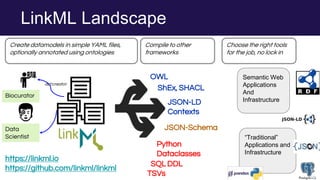
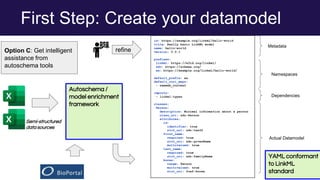
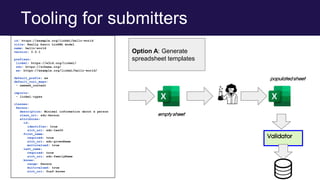
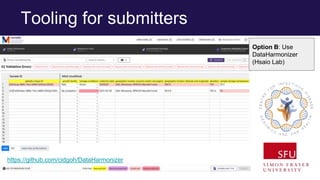
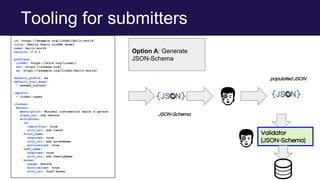
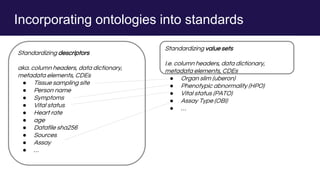
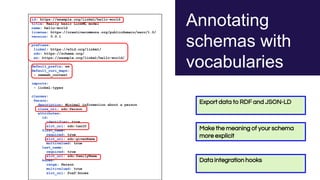
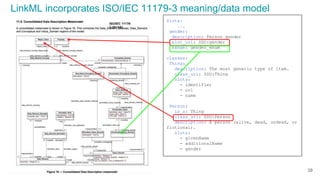
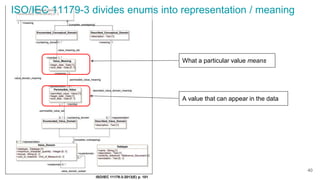
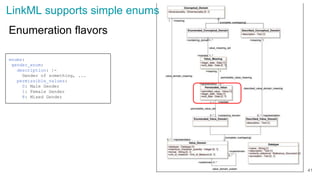
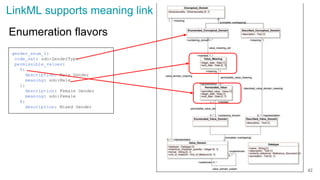
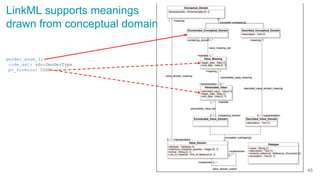
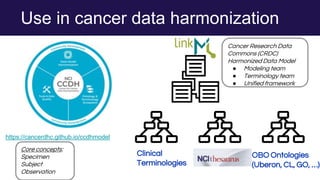
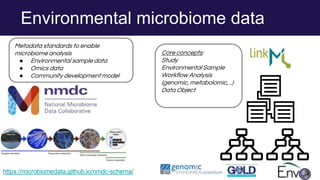
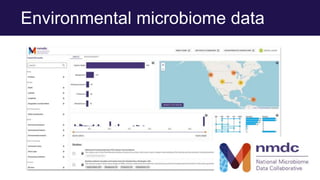
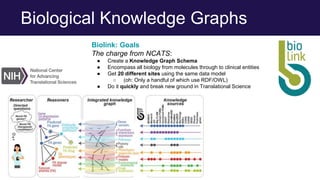
The document discusses the Linked Data Modeling Language (LinkML) as a framework for better structuring and integrating biomedical data, while highlighting the importance of ontologies and standardized vocabularies. It outlines the current challenges in data integration, including the underuse of ontologies and the prevalence of non-machine actionable standards. The presentation also emphasizes the need for effective tools and a universal framework to facilitate data standardization and improve interoperability in biomedical research.