More Related Content
Similar to PAG poster Jeevan Adhikari (20)
PAG poster Jeevan Adhikari
- 1. RESEARCH POSTER PRESENTATION DESIGN © 2012
www.PosterPresentations.com
The genetic base of modern cotton varieties is severely constricted
due to evolutionary and breeding bottlenecks imposed by
speciation, domestication and selection. Incorporation of
favorable alleles from exotic Gossypium accessions appears to be a
plausible approach to broaden the genetic base of cultivated
cotton. To analyze and evaluate the effects of exotic alleles on a
range of fiber quality traits across different genetic backgrounds
of cultivated cotton, we developed intraspecific populations from
crosses between an exotic/converted accession of upland cotton
(MDN101), that was prioritized for several important attributes,
and four elite cultivars that sample the gene pools of different US
production regions, studying the F2 and F3 generations for fiber
length, strength, fineness, elongation, uniformity and short fiber
content. We observed transgressive segregation for these traits,
supporting the hypothesis that the exotic donor contains novel
alleles of value for improving elite cottons, and providing strong
justification for genetic mapping to identify diagnostic DNA
markers. We revealed a significant positive relationship between
fiber length and strength, fiber length and uniformity index, and
fiber strength and uniformity index. Significant positive regression
coefficients in the F2-F3 generations for fiber elongation in the
MDN101×Acala Maxxa population, and for fiber length and strength
in the MDN101×PMHS200 population indicate relatively high
heritability of these traits and suitability of early-generation
populations for diagnostic marker identification. Genetic mapping
and expected validation of the presence of favorable transgressive
alleles would provide strong support for deeper exploration of
diversity within exotic G. hirsutum genotypes.
INTRODUCTION
To explore exotic/converted accessions of Upland cotton
(Gossypium hirsutum L.) with an aim to identify new alleles and
allele combinations useful in cotton improvement.
To evaluate the effects of exotic alleles and genetic backgrounds
on different fiber quality traits in Upland cotton.
RESEARCH OBJECTIVES
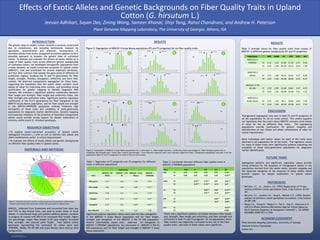
Figure 2: Segregation of MDN101 X Acala Maxxa populations (F2 and F3 progenies) for six fiber quality traits
RESULTS
Figure 2: Segregation of MDN101 X Acala Maxxa populations for a. Micronaire b. Fiber length (inches) c. Uniformity index (percentage) d. Fiber strength (grams/tex) e.
Elongation (percentage) and f. Short fiber content (percentage). Y axis indicates frequency and X axis indicates respective traits. Transgressive segregation was seen in all the
populations (both F2 and F3 progenies) for each of the six traits scored.
DISCUSSION
Transgressive segregation was seen in both F2 and F3 progenies of
all the populations for all six traits scored. This clearly supports
our hypothesis that the exotic donor MDN101 contains novel alleles
of value for the six different fiber traits. The segregating
populations provide justification for genetic mapping and
identification of new alleles and allele combinations of value for
cotton improvement.
Many individuals with better values for each of the traits were
observed in all populations for all the traits. The regression values
for many of these traits were significantly positive indicating the
suitability of these early-generation populations for diagnostic
marker identification.
FUTURE TASKS
• McCarty J.C., Jr., Jenkins J.N. (1993) Registration of 79 day-
neutral primitive cotton germplasm lines. Crop Science 33:351-
351.
• McCarty J.C., Jenkins J.N., Tang B., Watson C.E. (1996) Genetic
analysis of primitive cotton germplasm accessions. Crop Science
36:581-585.
• Wang Z.N., Zhang D., Wang X.Y., Tan X., Guo H., Paterson A.H.
(2013) A Whole-Genome DNA Marker Map for Cotton Based on
the D-Genome Sequence of Gossypium raimondii L. G3-GENES
GENOMES GENETICS 3:1759.
Plant Genome Mapping Laboratory, The University of Georgia, Athens, GA
Jeevan Adhikari, Sayan Das, Zining Wang, Sameer Khanal, Shiyi Tang, Rahul Chandnani, and Andrew H. Paterson
Effects of Exotic Alleles and Genetic Backgrounds on Fiber Quality Traits in Upland
Cotton (G. hirsutum L.)
MATERIALS AND METHODS
MDN101, collected from Guatemala and converted from short day
line T101 to day-neutral form, was used as exotic donor of novel
alleles. It contributed large and positive additive genetic variation
to progeny of crosses with DPL16 for increased fiber length, higher
lint percentage, longer fiber length (2.5% span length), increased
fiber strength, and reduced plant height. Four Upland cotton
genotypes representing the cultivated US cotton gene pool
(PD94042, DES56, PM HS 200 and Acala Maxxa) were used as elite
backgrounds.
0
5
10
15
20
25
0.89
0.91
0.93
0.95
0.97
0.99
1.01
1.03
1.05
1.07
1.09
1.11
1.13
1.15
1.17
1.19
1.21
1.23
1.25
1.27
1.29
Acala MaxxaMDN101
F2
0
2
4
6
8
10
12
14
16
18
20
0.89
0.91
0.93
0.95
0.97
0.99
1.01
1.03
1.05
1.07
1.09
1.11
1.13
1.15
1.17
1.19
1.21
1.23
1.25
1.27
1.29
Acala MaxxaMDN101
F3
0
2
4
6
8
10
12
14
16
18
77
78
78
79
79
80
80
81
81
82
82
83
83
84
84
85
85
86
86
87
MDN 101 Acala Maxxa
F2
0
5
10
15
20
25
77
78
78
79
79
80
80
81
81
82
82
83
83
84
84
85
85
86
86
87
MDN 101
Acala Maxxa
F3
0
5
10
15
20
25
30
3.3
3.5
3.8
4.0
4.3
4.5
4.8
5.0
5.3
5.5
5.8
6.0
6.3
6.5
6.8
7.0
7.3
7.5
7.8
8.0
8.3
8.5
Acala Maxxa MDN 101
F2
0
2
4
6
8
10
12
14
16
18
3.3
3.5
3.8
4.0
4.3
4.5
4.8
5.0
5.3
5.5
5.8
6.0
6.3
6.5
6.8
7.0
7.3
7.5
7.8
8.0
8.3
8.5
Acala Maxxa MDN 101
F3
0
5
10
15
20
25
23
24.5
26
27.5
29
30.5
32
33.5
35
36.5
Acala Maxxa
F2
MDN101
0
5
10
15
20
25
30
35
23
24.5
26
27.5
29
30.5
32
33.5
35
36.5
MDN101 Acala
maxxa
F3
0
5
10
15
20
25
6.0
6.4
6.8
7.2
7.6
8.0
8.4
8.8
9.2
9.6
10.0
10.4
10.8
11.2
11.6
12.0
12.4
12.8
13.2
13.6
Acala Maxxa MDN 101
F2
0
5
10
15
20
25
30
35
40
6.0
6.4
6.8
7.2
7.6
8.0
8.4
8.8
9.2
9.6
10.0
10.4
10.8
11.2
11.6
12.0
12.4
12.8
13.2
13.6
Acala
Maxxa
MDN 101
F3
fed
b c
Populations
Traits
MDN101 ×PD
94042
MDN101 × DES
56
MDN101 × Acala
Maxxa
MDN101 ×
PMHS200
Micronaire 0.35* -0.03 0.28 0.36*
Fiber length 0.06 0.08 0.48* 0.75**
Uniformity 0.09 0.23 0.11 0.33*
Strength 0.12 -0.11 0.46* 0.41*
Elongation 0.34* 0.04 0.68** 0.61**
Short fiber(%) 0.14 -0.02 0.14 0.08
Table 1: Regression of F3 progenies over F2 progenies for different
traits in different populations
Table 2: Correlation between different fiber quality traits in
MDN101 X PD94042 populations
Traits UHM UI STR ELO SFC
MIC F2 0.03 0.23 0.05 0.33 -0.41*
F3 0.08 0.32 0.28 0.18 -0.59**
UHM F2 0.59** 0.73** 0.27 -0.56**
F3 0.74** 0.74** -0.31 -0.4*
UI F2 0.63** 0.45* -0.74**
F3 0.67** -0.12 -0.62**
STR F2 0.62** -0.69**
F3 -0.37 -0.49*
ELO F2 -0.65**
F3 0.003
Figure 1: Diversity of parents used for creating experimental populations. Green arrow
indicates exotic parent and red arrows indicate elite genotypes of Upland cotton.
Significant positive regression values were seen for fiber elongation
in the MDN101 X Acala Maxxa population and for fiber length,
strength and elongation in the MDN101 X PM HS 200 population.
Positive intermediate values were observed for elongation in
MDN101 X PD94042 populations, for micronaire in MDN101 X PM HS
200 populations and for fiber length and strength in MDN101 X Acala
Maxxa population.
There was a significant positive correlation between fiber length
and strength, fiber length and uniformity, and fiber strength and
uniformity in both F2 and F3 progenies for all populations. Short
fiber content was negatively correlated to the rest of the fiber
quality traits and most of these values were significant .
0
2
4
6
8
10
12
14
16 MDN 101
Acala Maxxa
F2
0
2
4
6
8
10
12
14
16
18
2.5
2.7
2.9
3.1
3.3
3.5
3.7
3.9
4.1
4.3
4.5
4.7
4.9
5.1
5.3
5.5
5.7
5.9
6.1
6.3
6.5
MDN 101Acala Maxxa
F3
a
REFERENCES
Segregation patterns and significant regression values provide
strong evidence for the presence of transgressive alleles in the
populations derived from the exotic donor. Genetic mapping and
the expected validation of the presence of these alleles would
provide support for deeper exploration of upland cotton
genotypes.
Population MIC UHM UI STR ELO SFC
MDN 101 X
PD94042 F2 5.25 1.02 81.84 27.39 4.37 8.44
F3 4.54 1.04 82.84 28.53 6.69 7.46
MDN 101 X
DES56 F2 5.37 1.00 81.07 26.41 4.37 8.99
F3 3.87 1.09 82.28 28.92 5.54 8.27
MDN101 X PM
HS 200 F2 5.20 0.99 81.08 28.81 3.97 8.72
F3 4.93 1.06 83.09 29.68 5.61 7.22
MDN 101 X
Acala maxxa F2 5.31 1.03 82.1 30.42 4.84 7.92
F3 4.29 1.09 83.39 30.48 6.31 7.13
Table 3: Average values for fiber quality traits from crosses of
MDN101 in different genetic backgrounds (F2 and F3 progenies)
RESULTS
ACKNOWLEDGEMENT
Plant Genome Mapping Laboratory, University of Georgia
National Science Foundation
Cotton Inc.