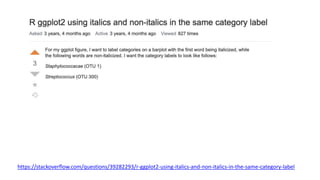
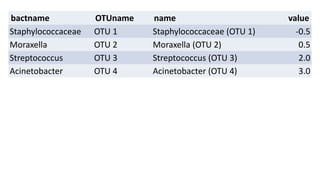
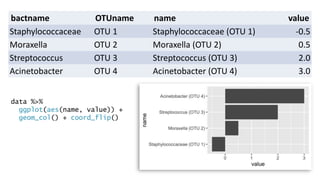
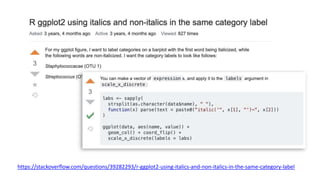
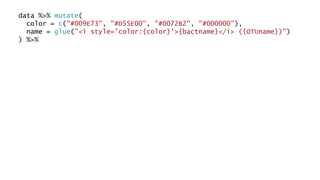
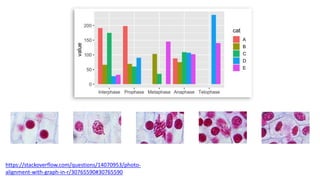
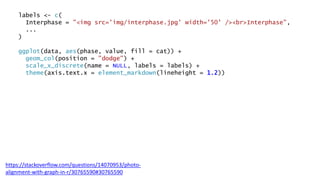
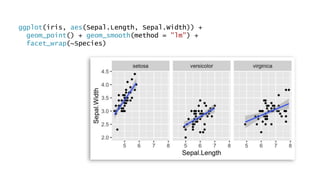
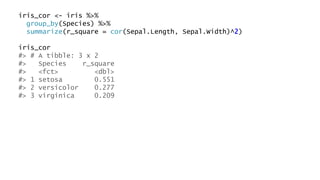
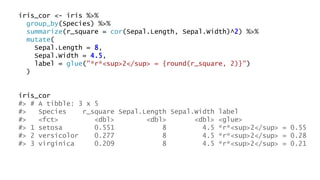
The document discusses how to add formatted text like italics, bold, colors and images to ggplot2 visualizations using the ggtext and gridtext packages. It provides examples of labeling bars and points with formatted text, adding correlation values alongside a scatter plot faceted by species, and controlling font properties and alignments. The key package that enables rich text formatting in plots is gridtext.