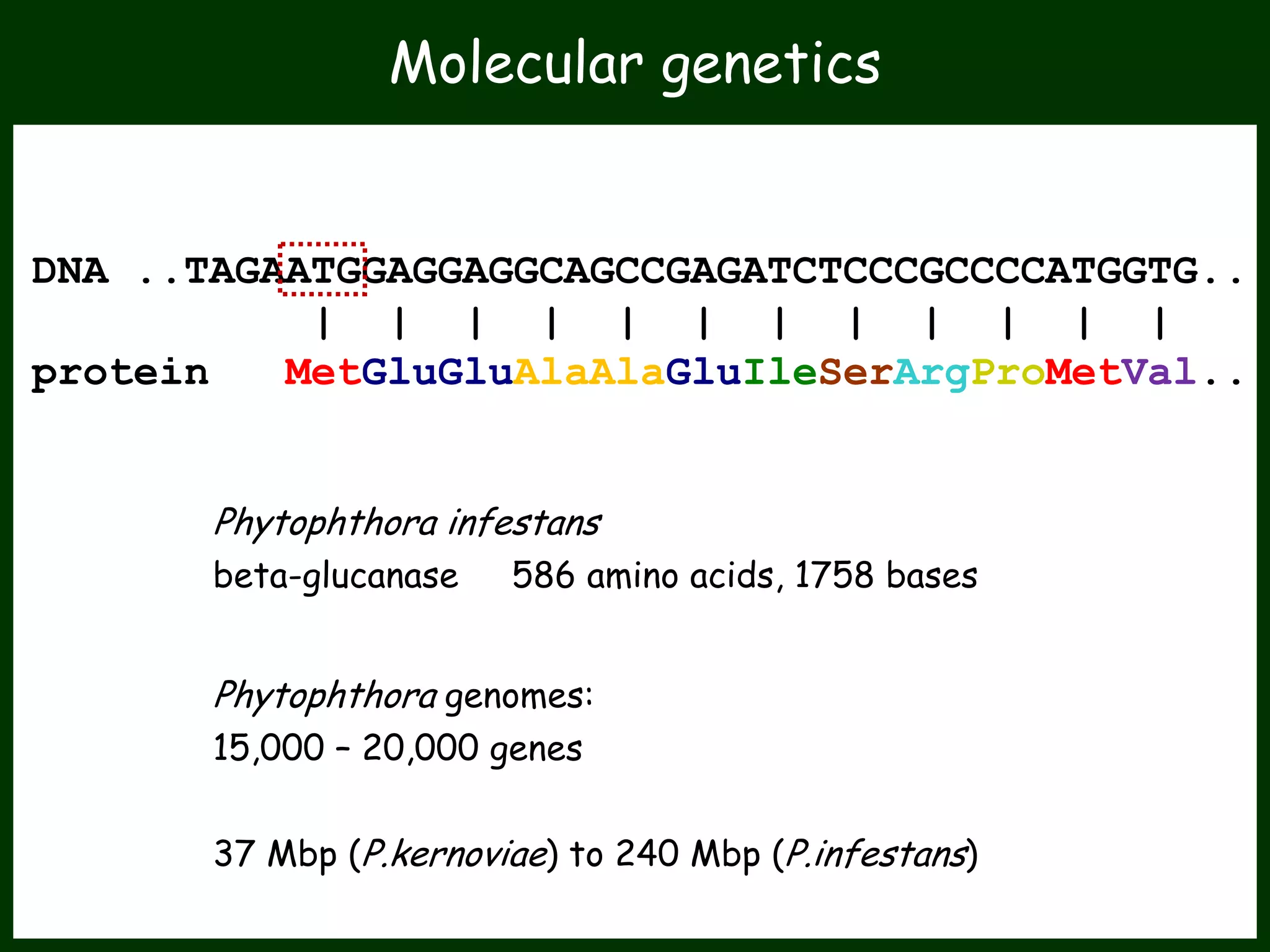
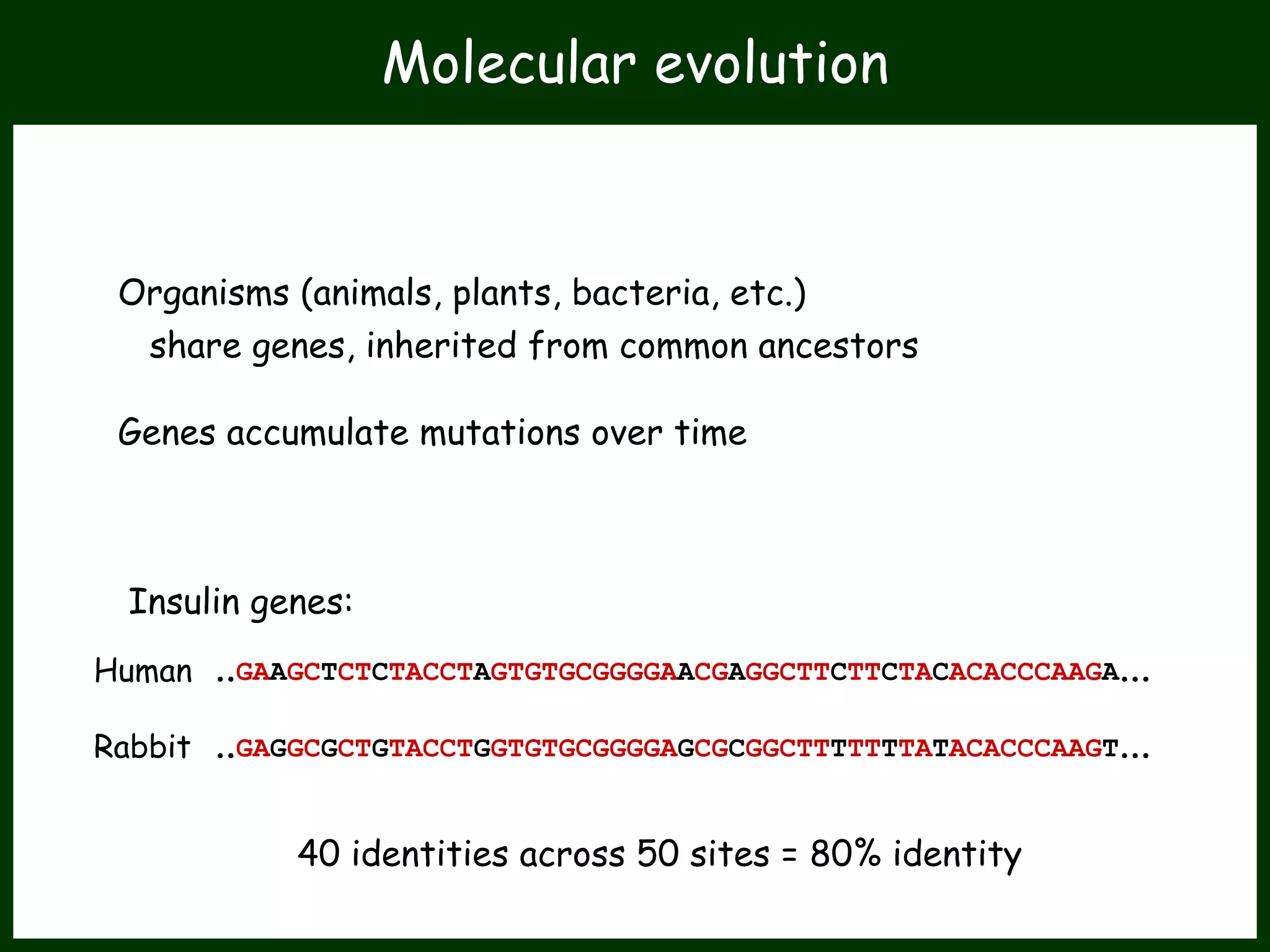
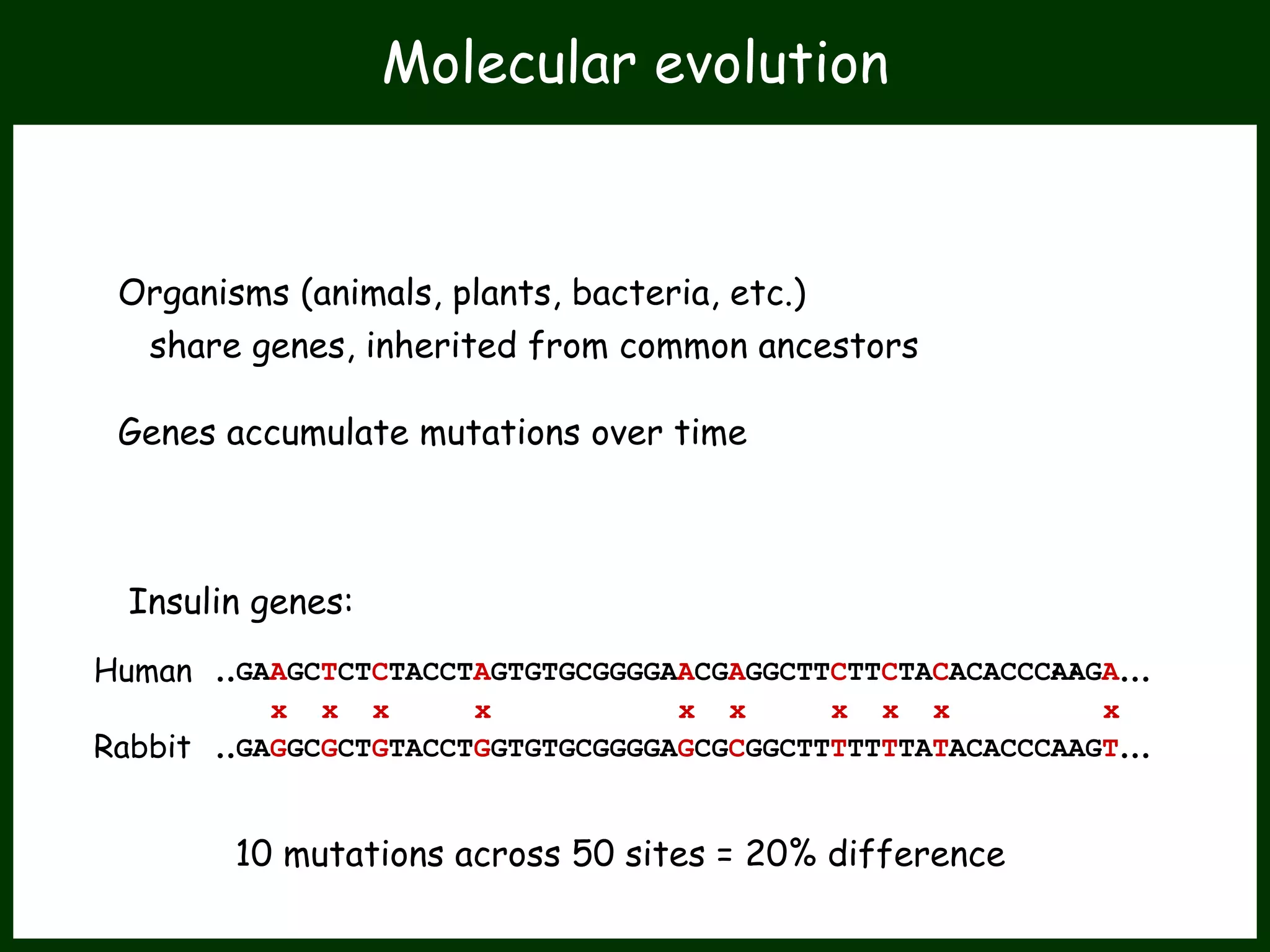
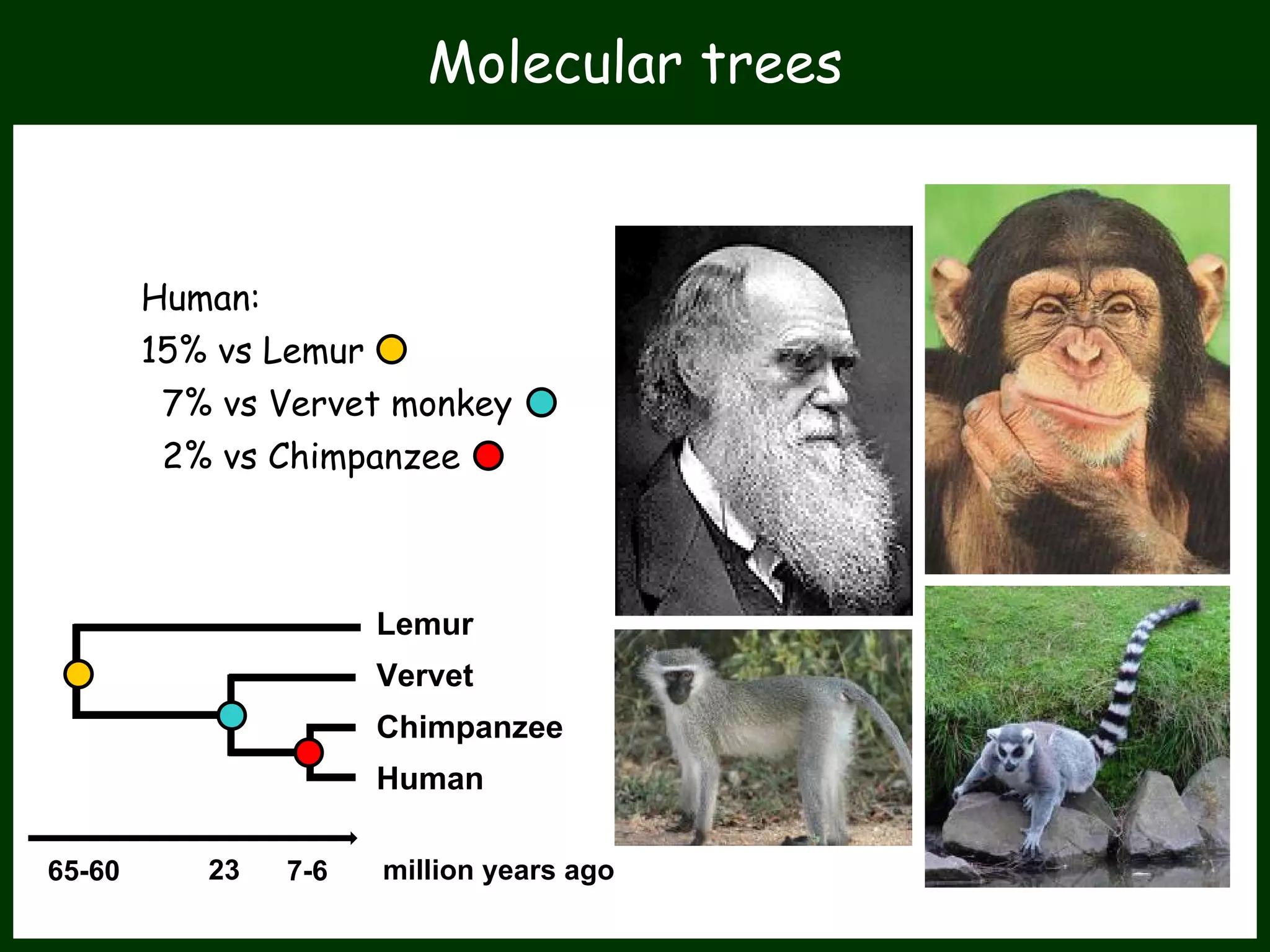
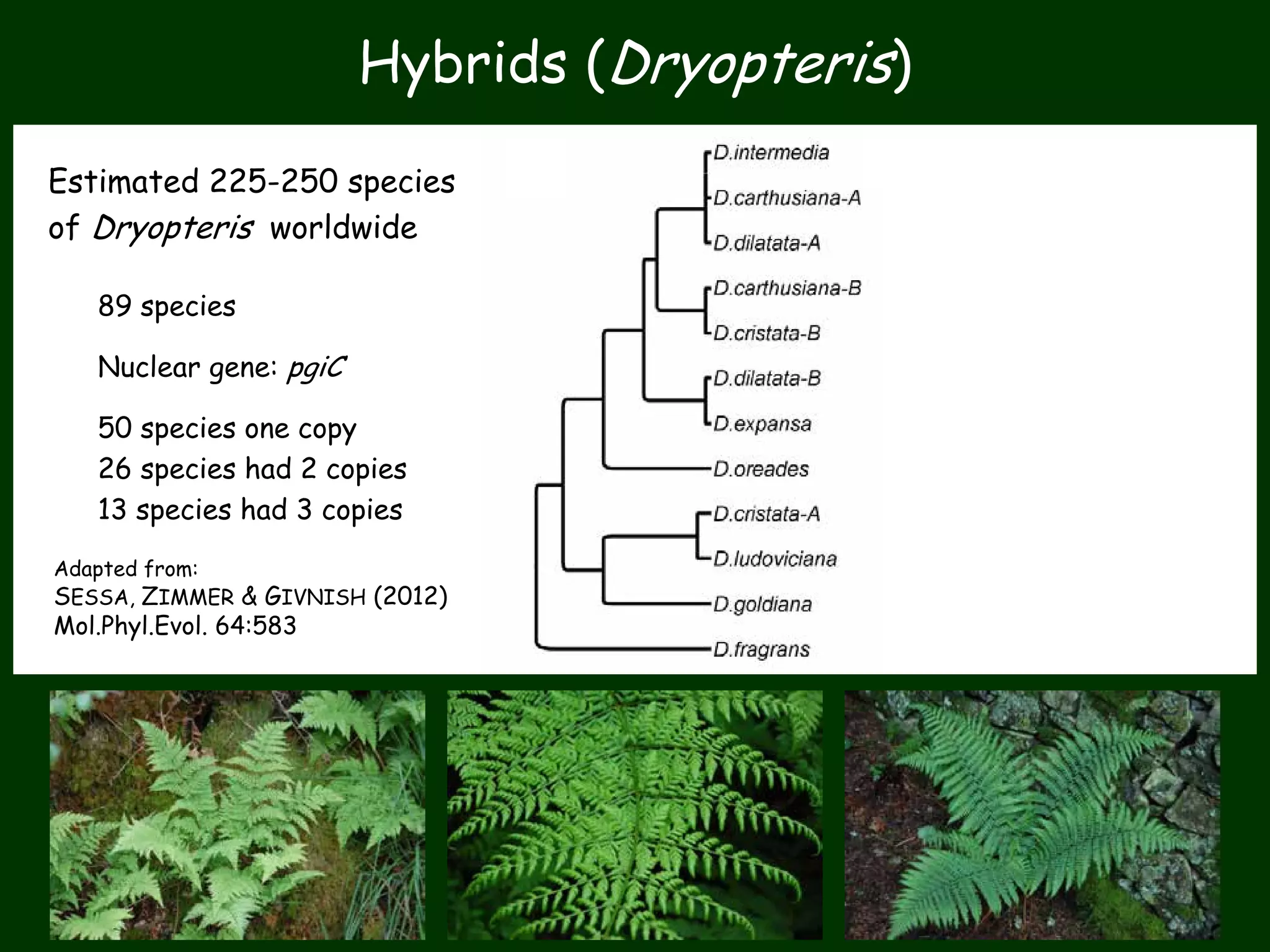
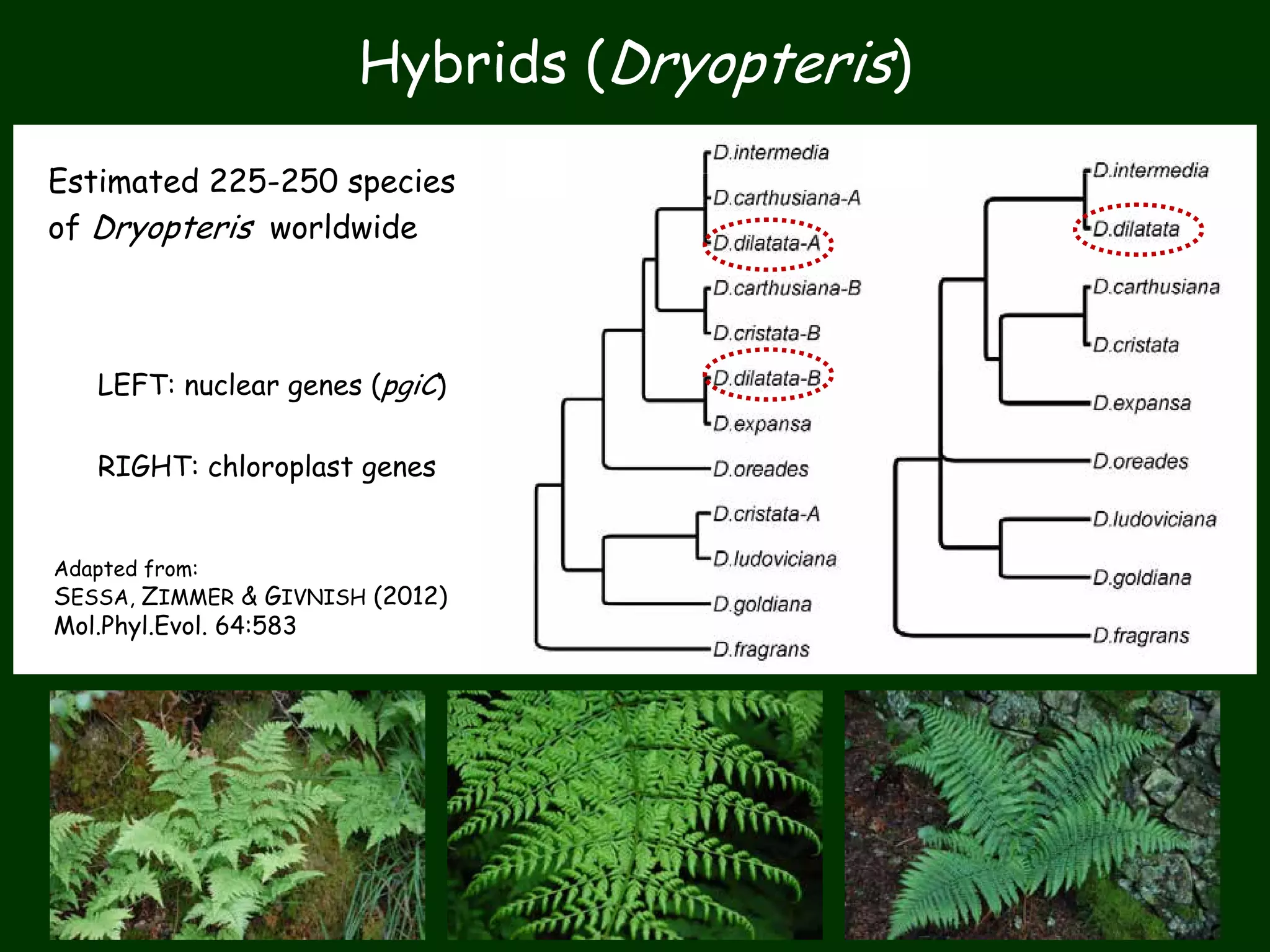
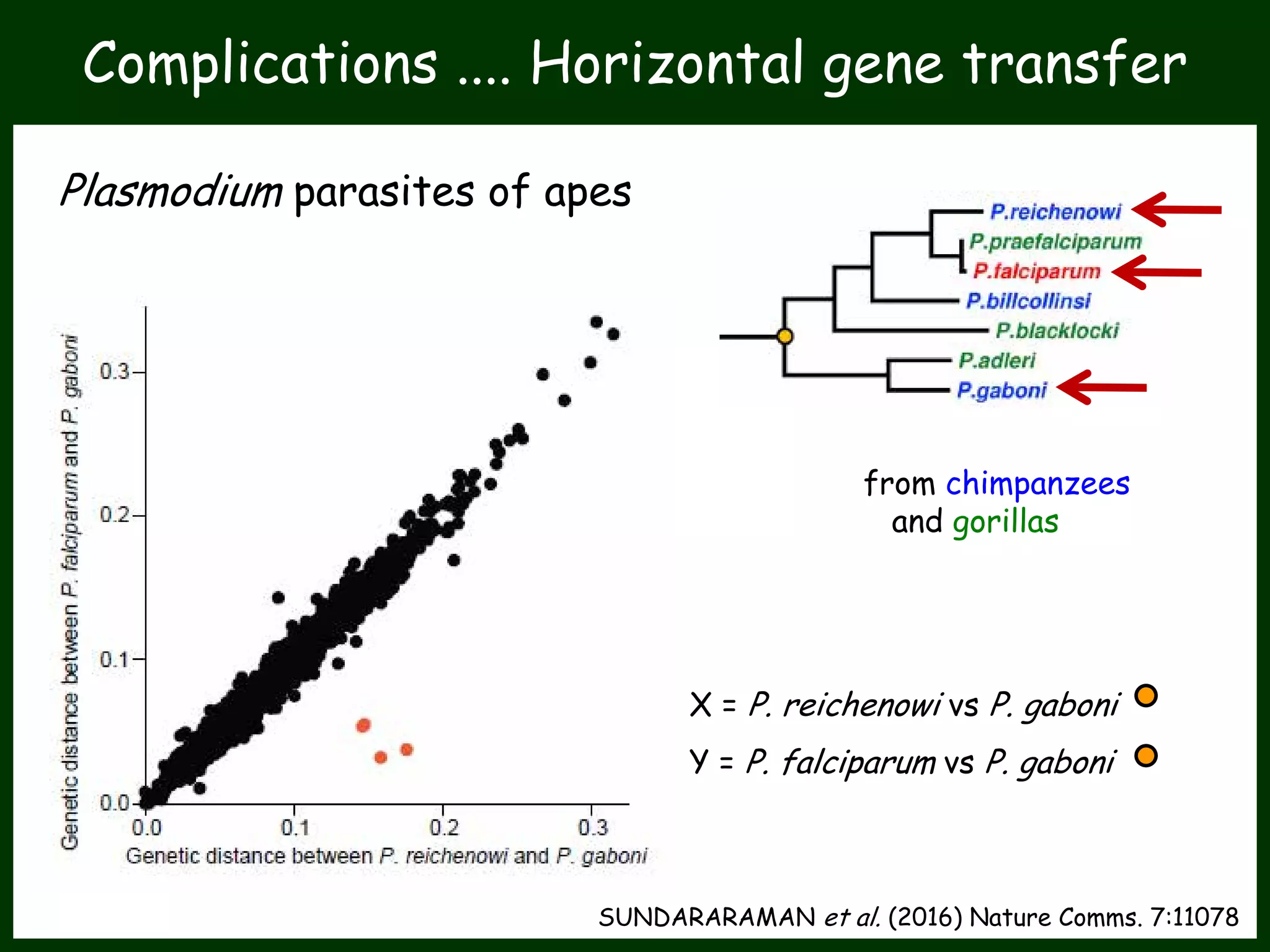
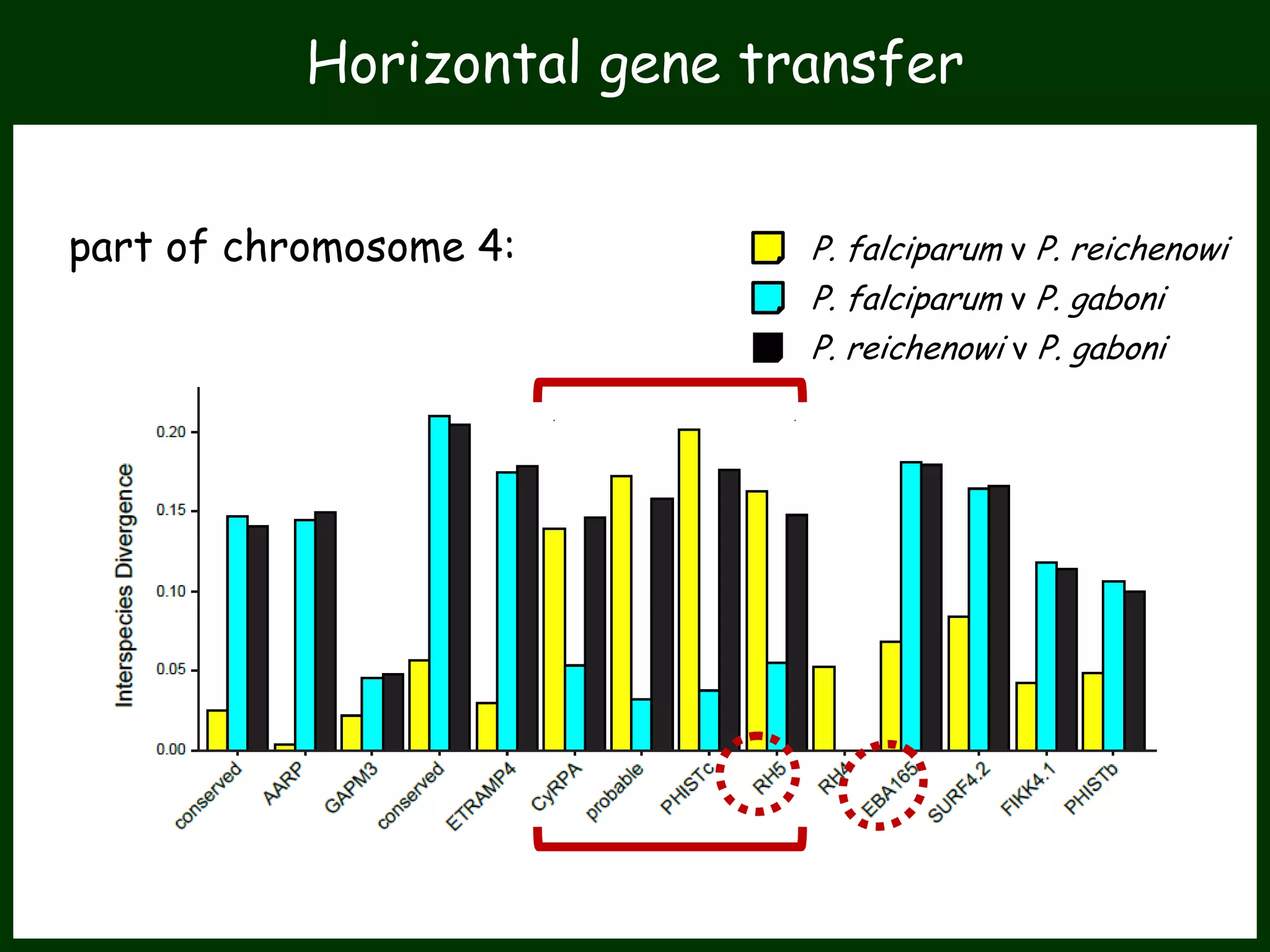
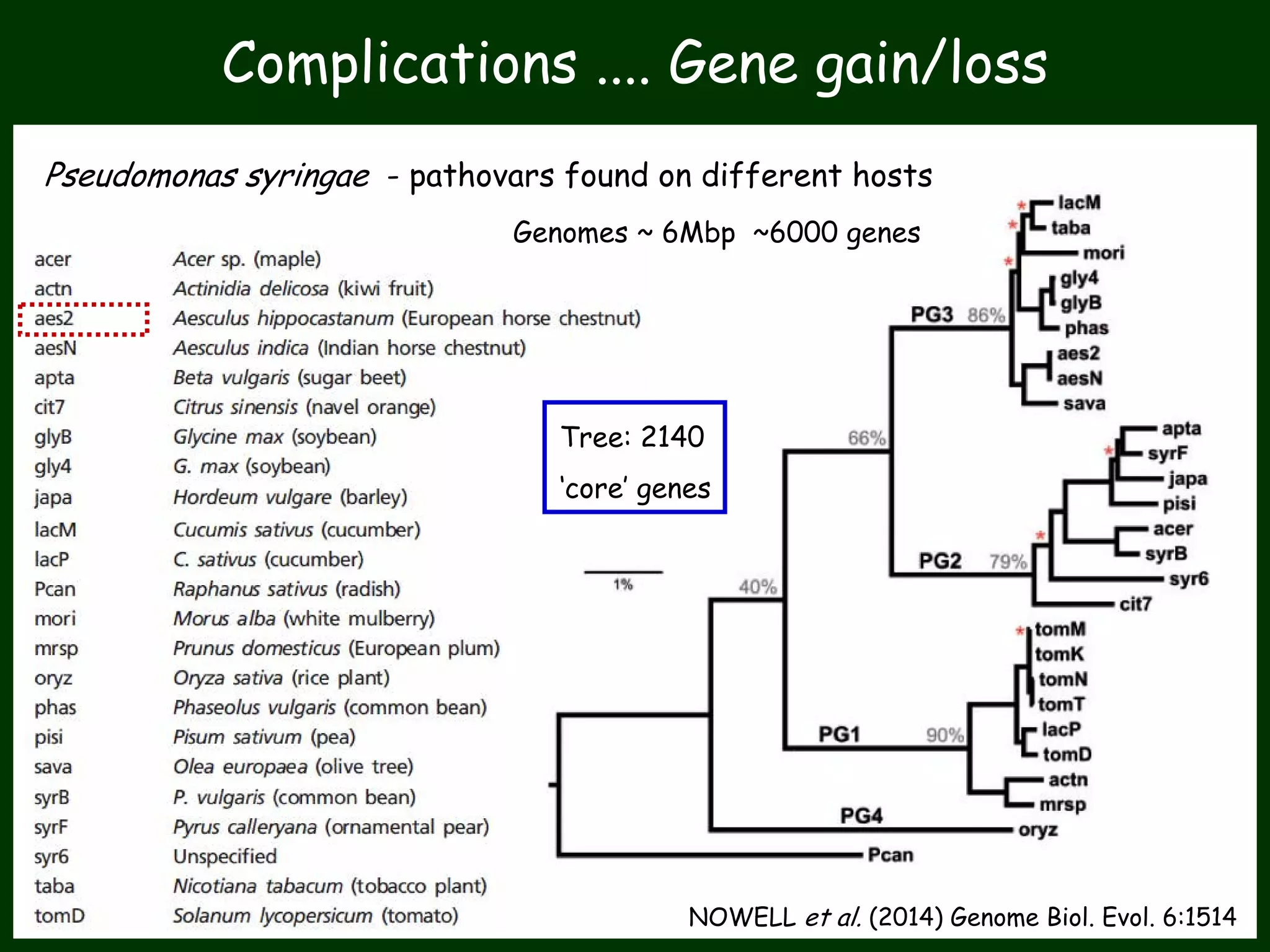
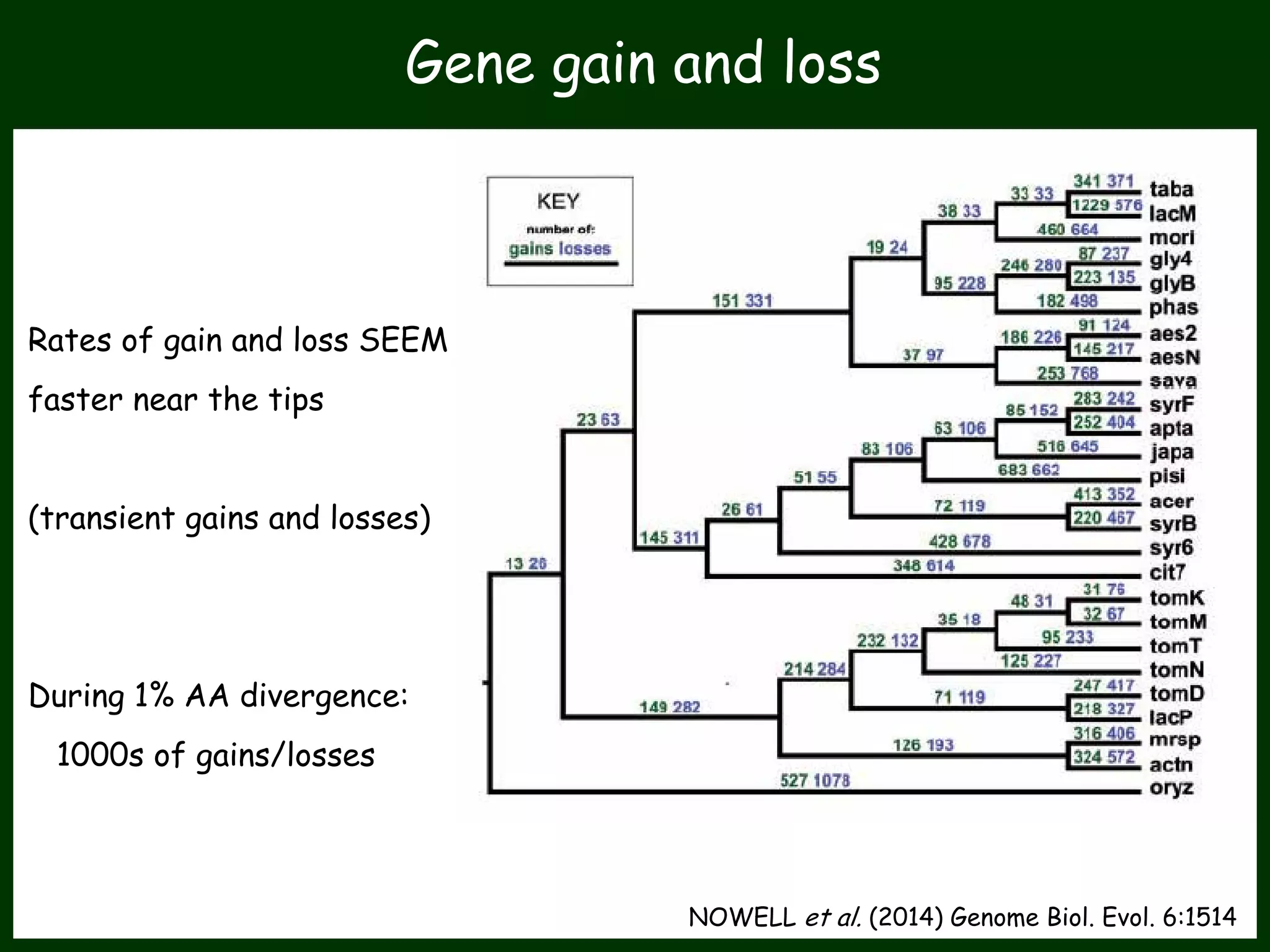
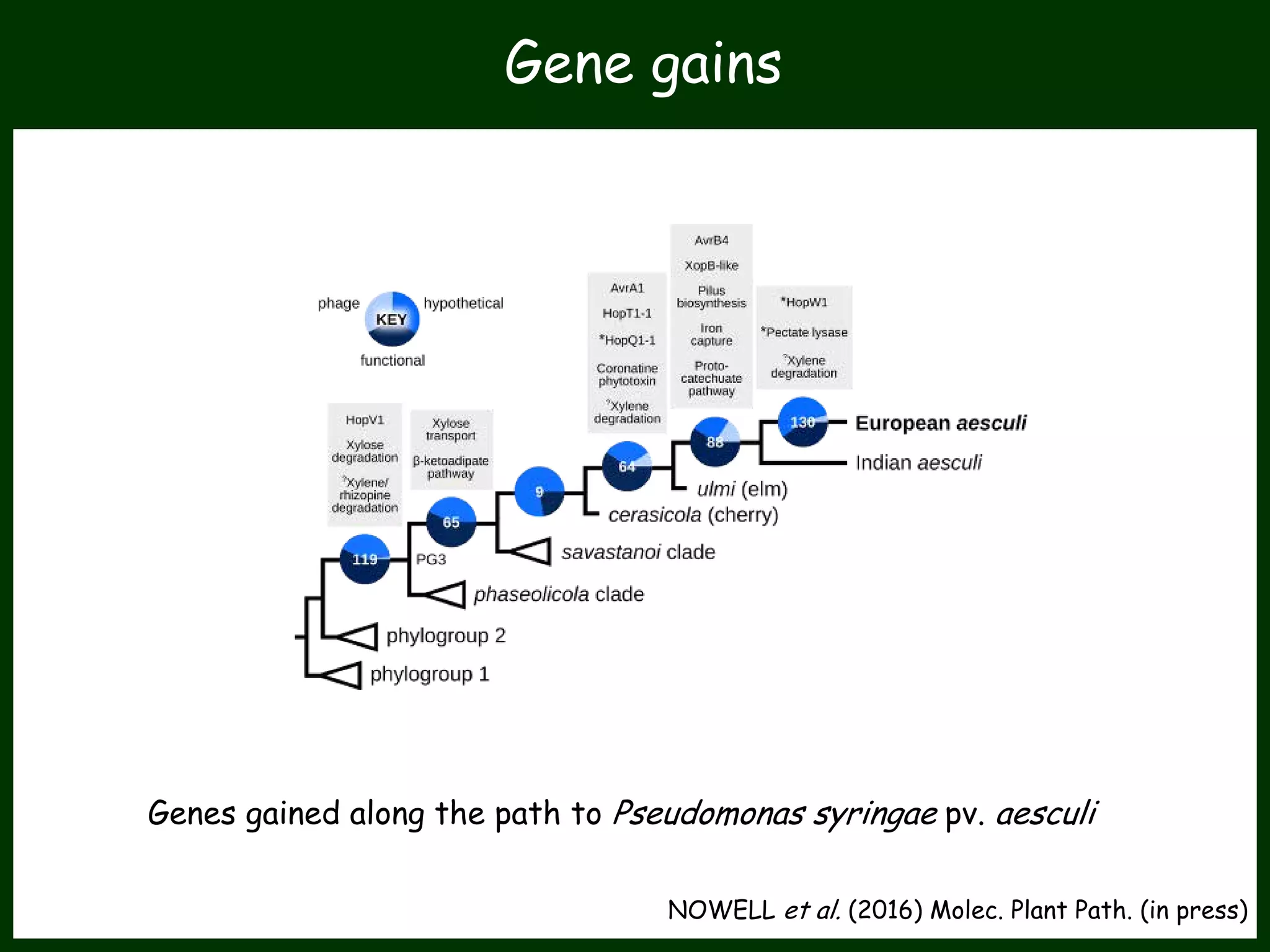
This work package aims to understand genome evolution in Phytophthora by analyzing UK diversity, origins of emerging strains, and mechanisms of adaptation such as hybridization and horizontal gene transfer. The researchers will sequence genomes to understand how Phytophthora adapts to new hosts, woody hosts, and the role of hybridization and gene transfer in evolution.