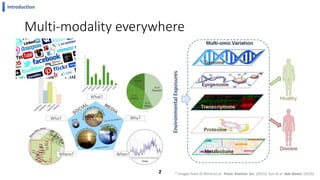
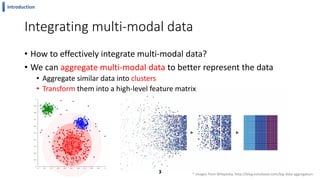
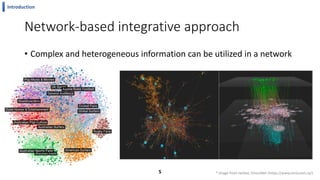
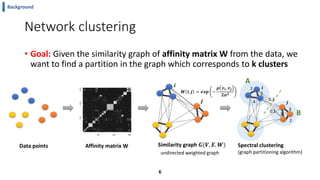
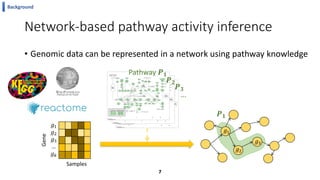
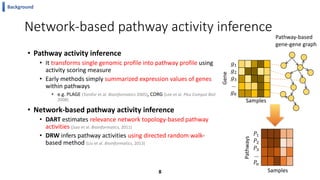
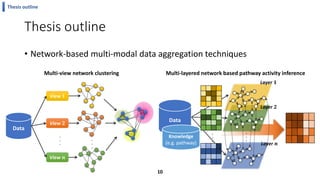
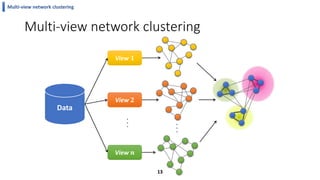
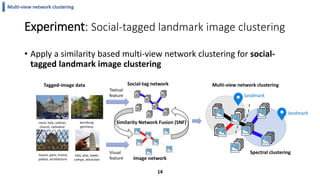
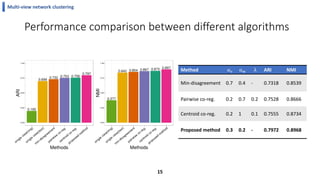
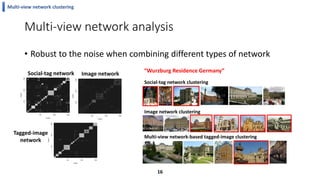
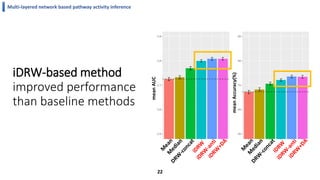
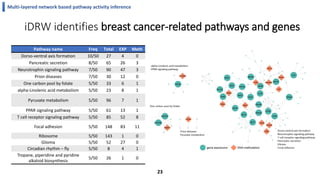
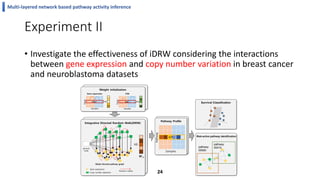
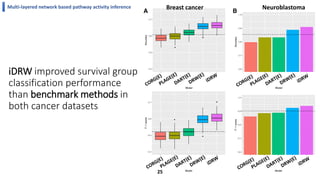
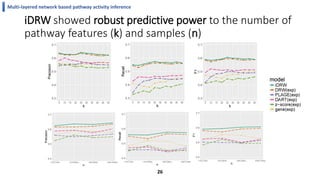
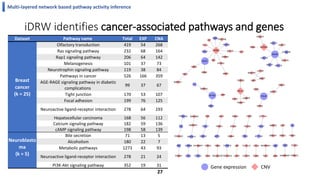
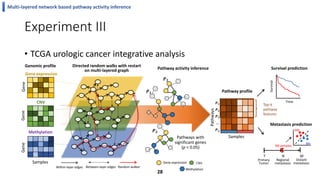
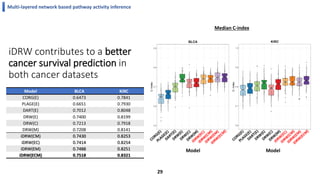
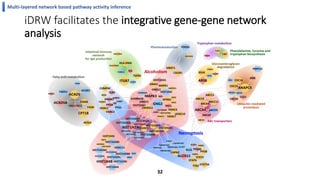
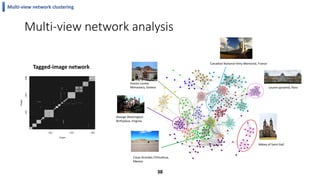
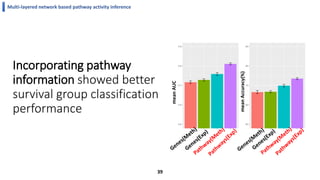
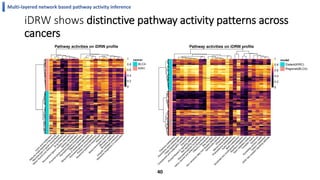
This document summarizes the author's PhD dissertation which develops network-based machine learning approaches for aggregating multi-modal data. It presents two key techniques: 1) multi-view network clustering to integrate different network views for clustering tasks like social-tagged image clustering, and 2) multi-layered network based pathway activity inference to infer pathway activities from integrated gene networks using techniques like directed random walks. It outlines experiments applying these techniques to various biomedical datasets for clustering, survival/metastasis prediction, and identifying cancer-related pathways and genes.