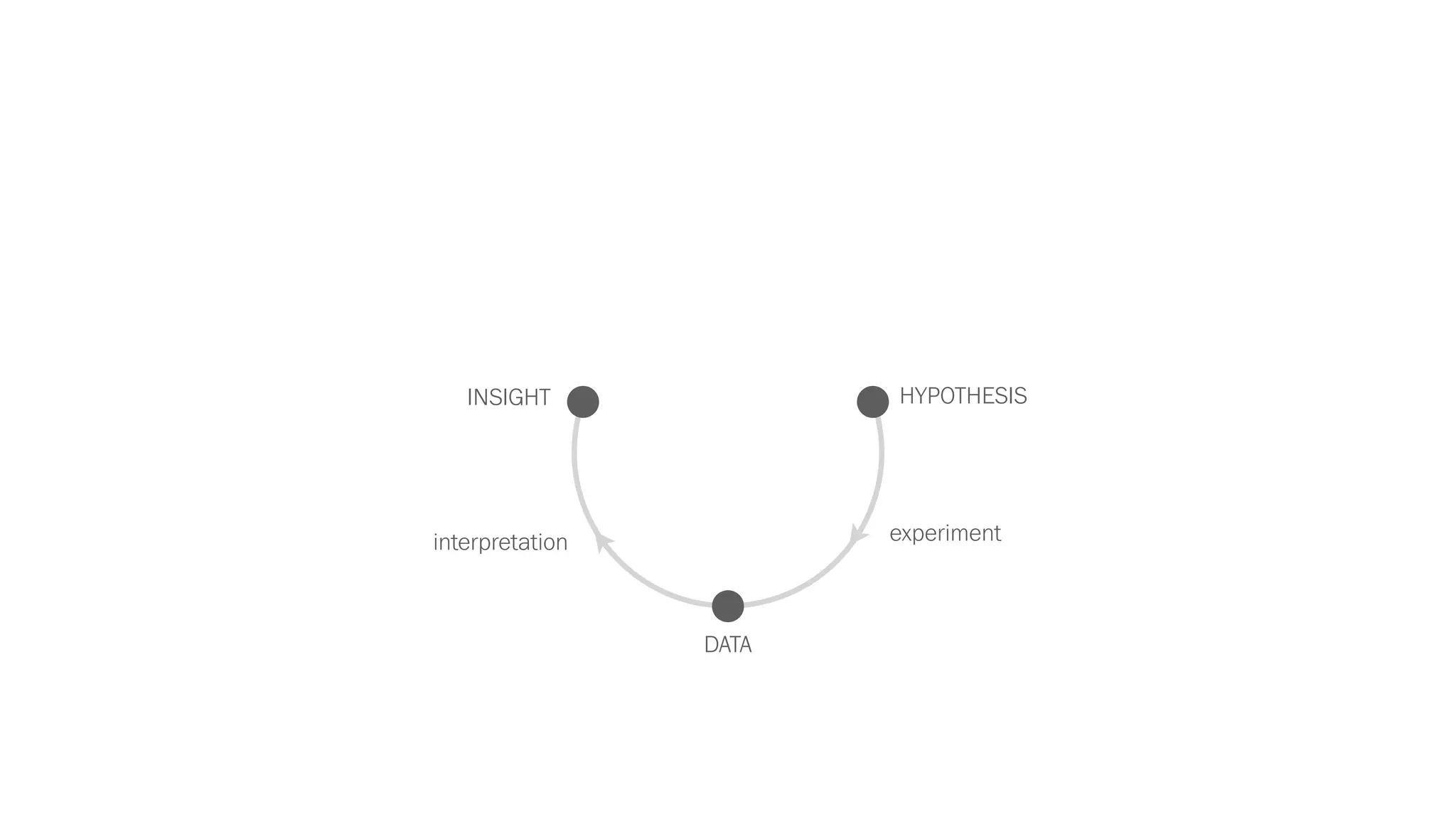
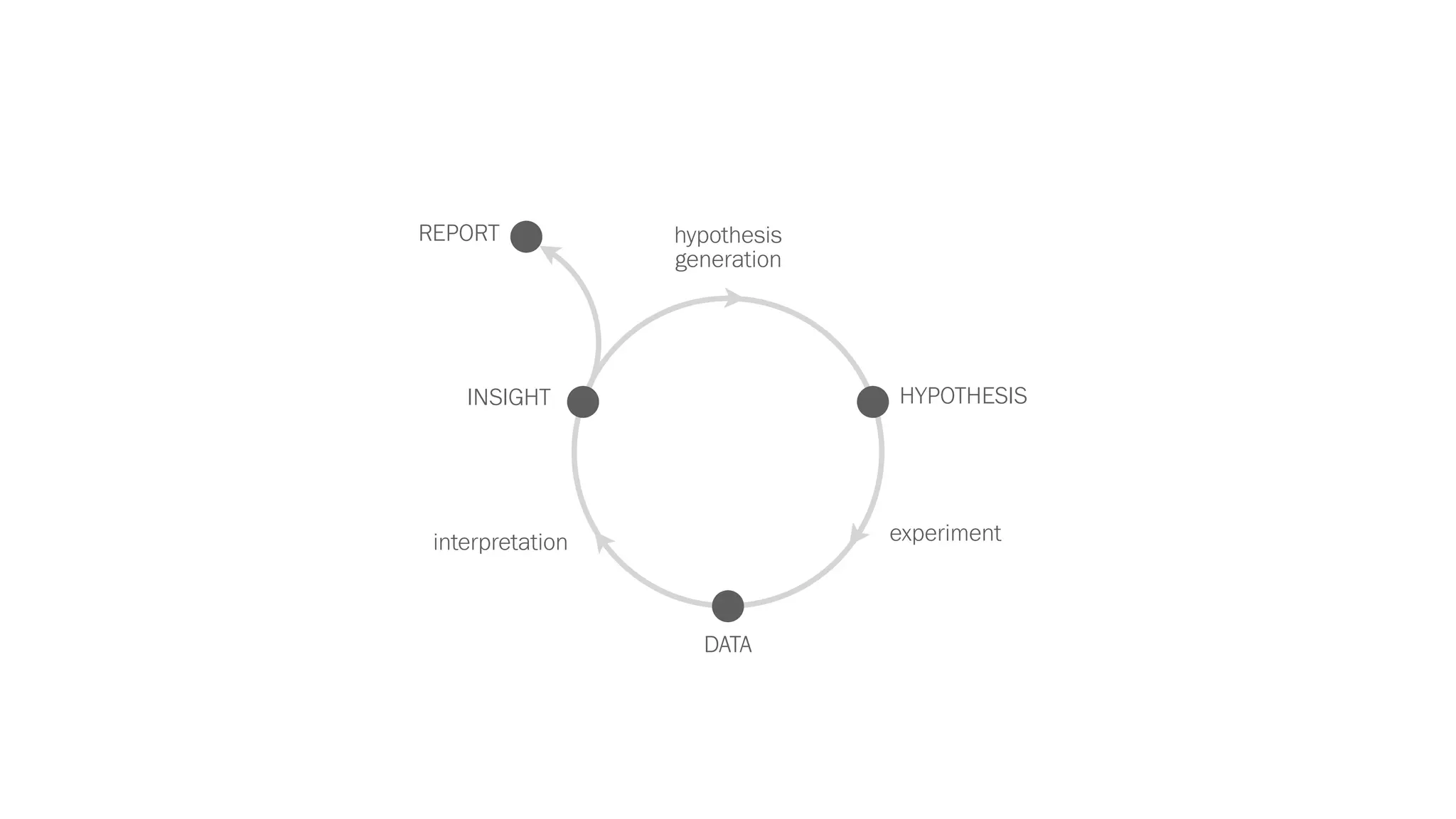
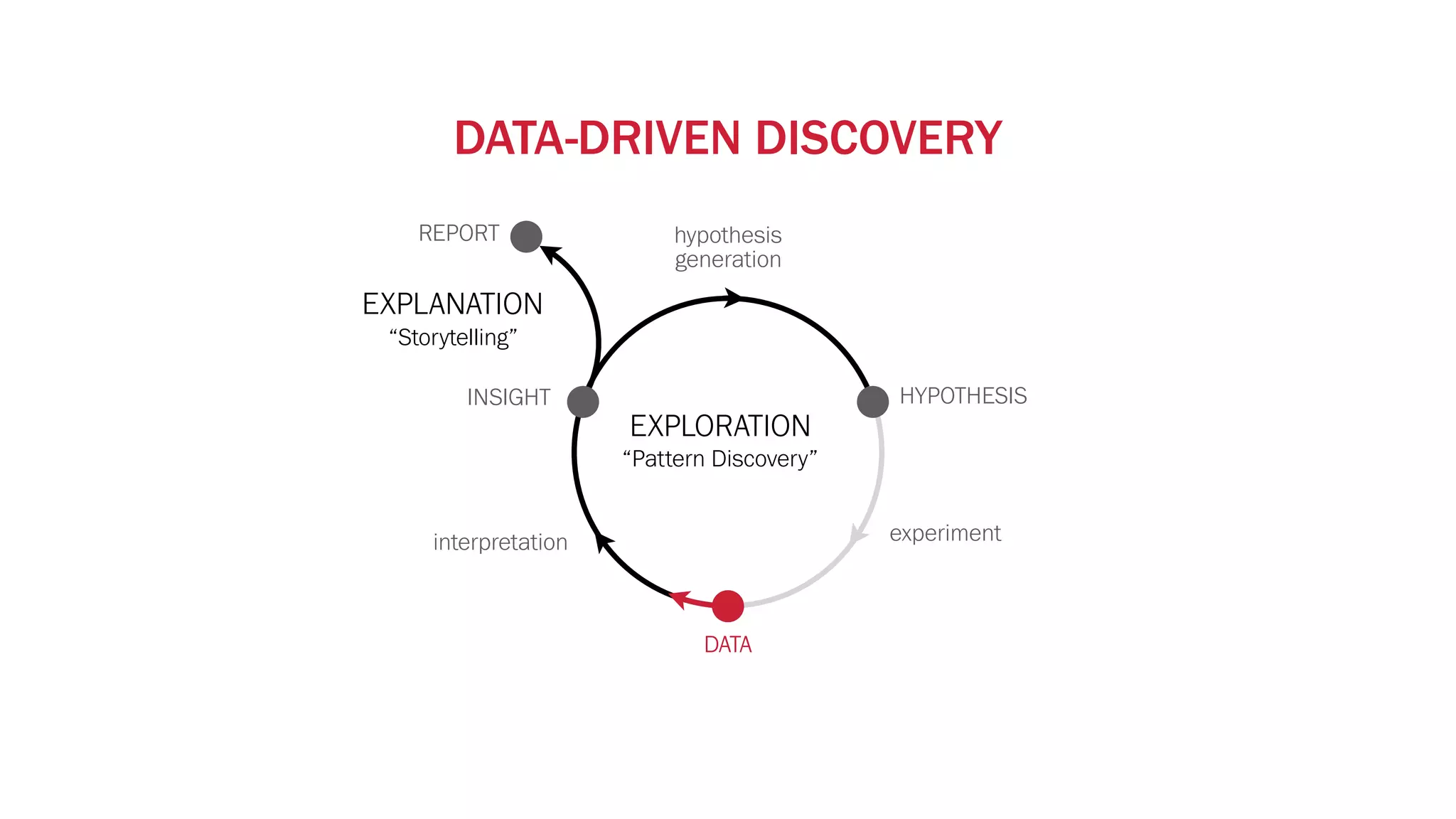
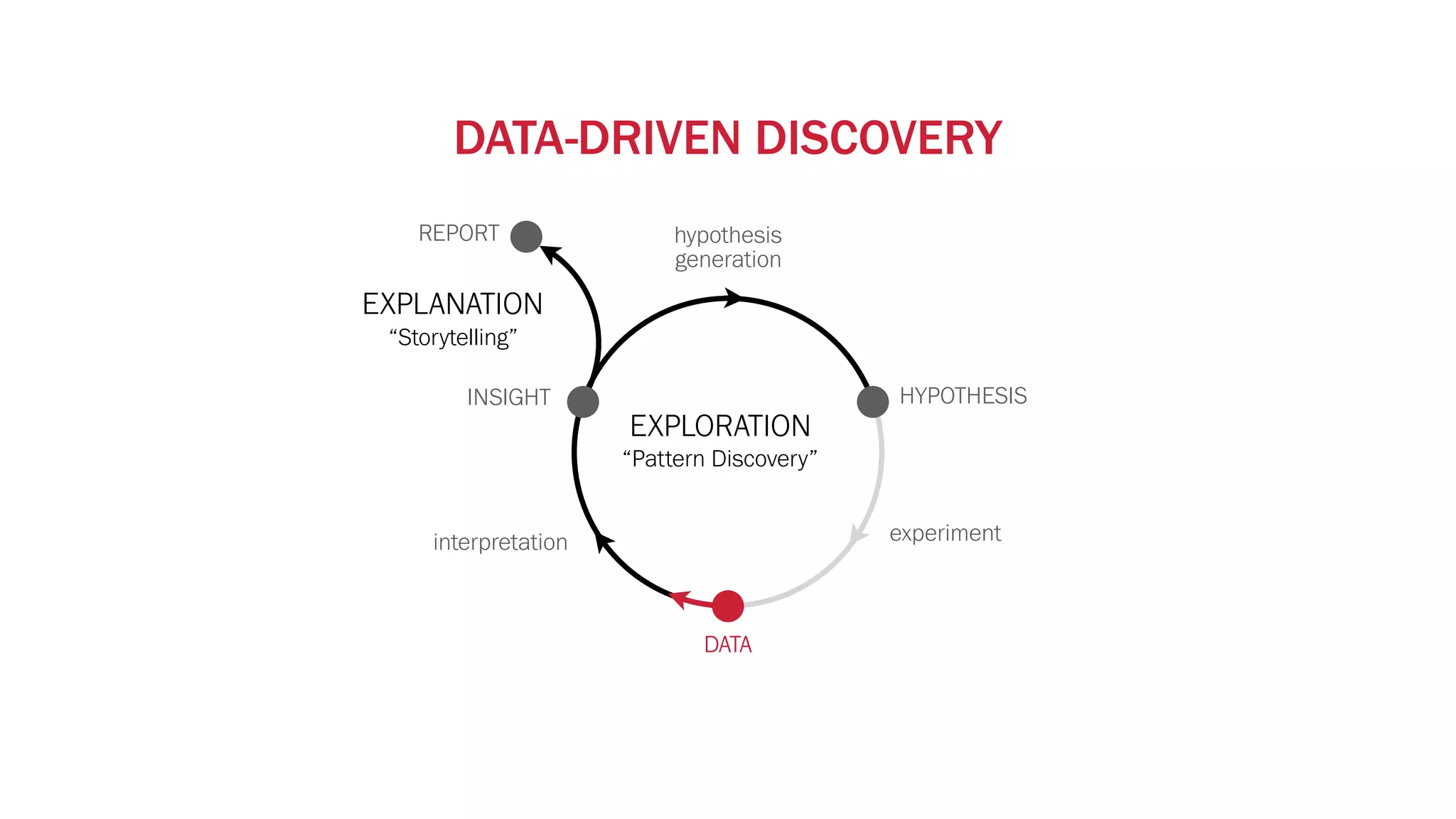
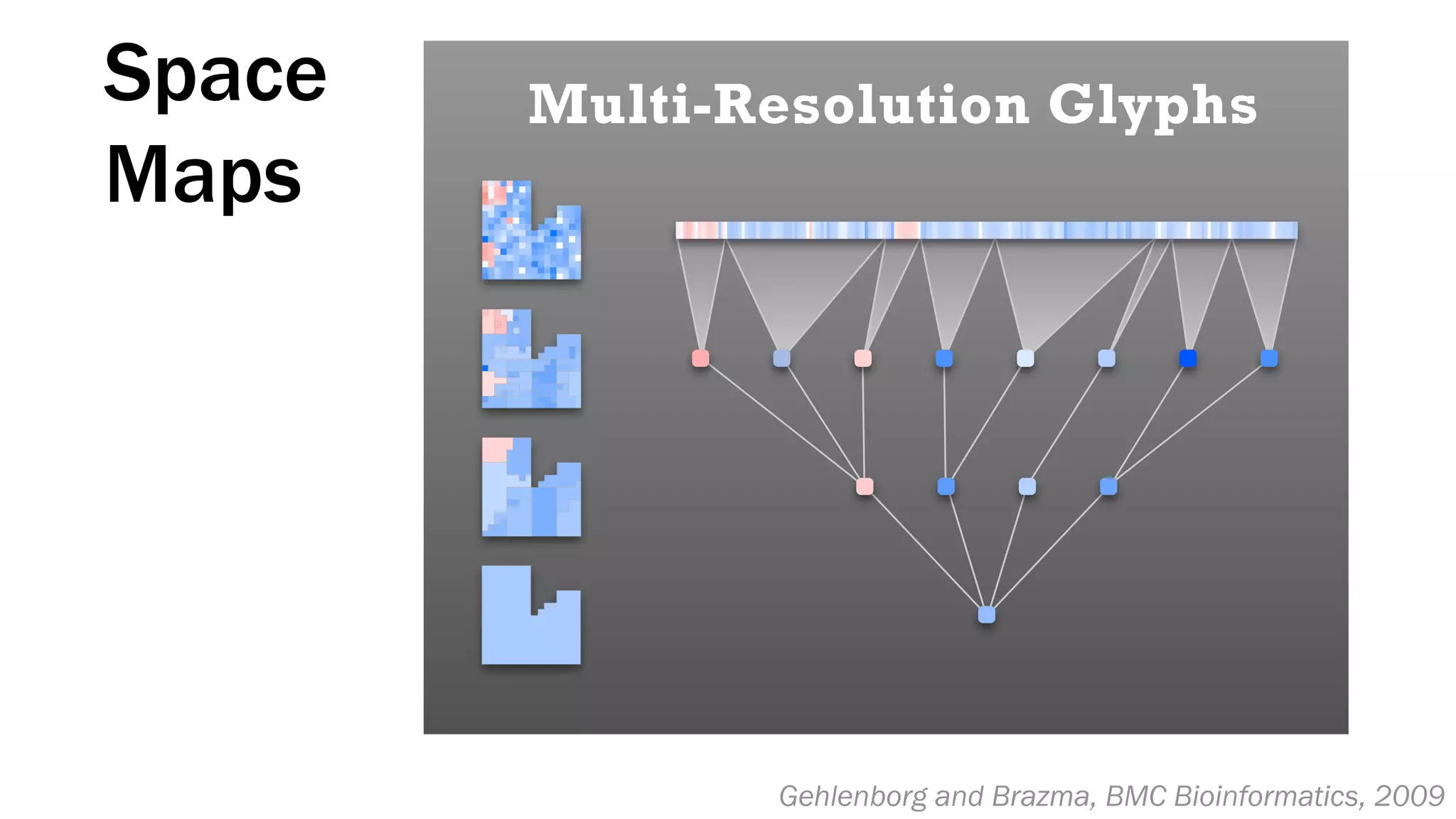
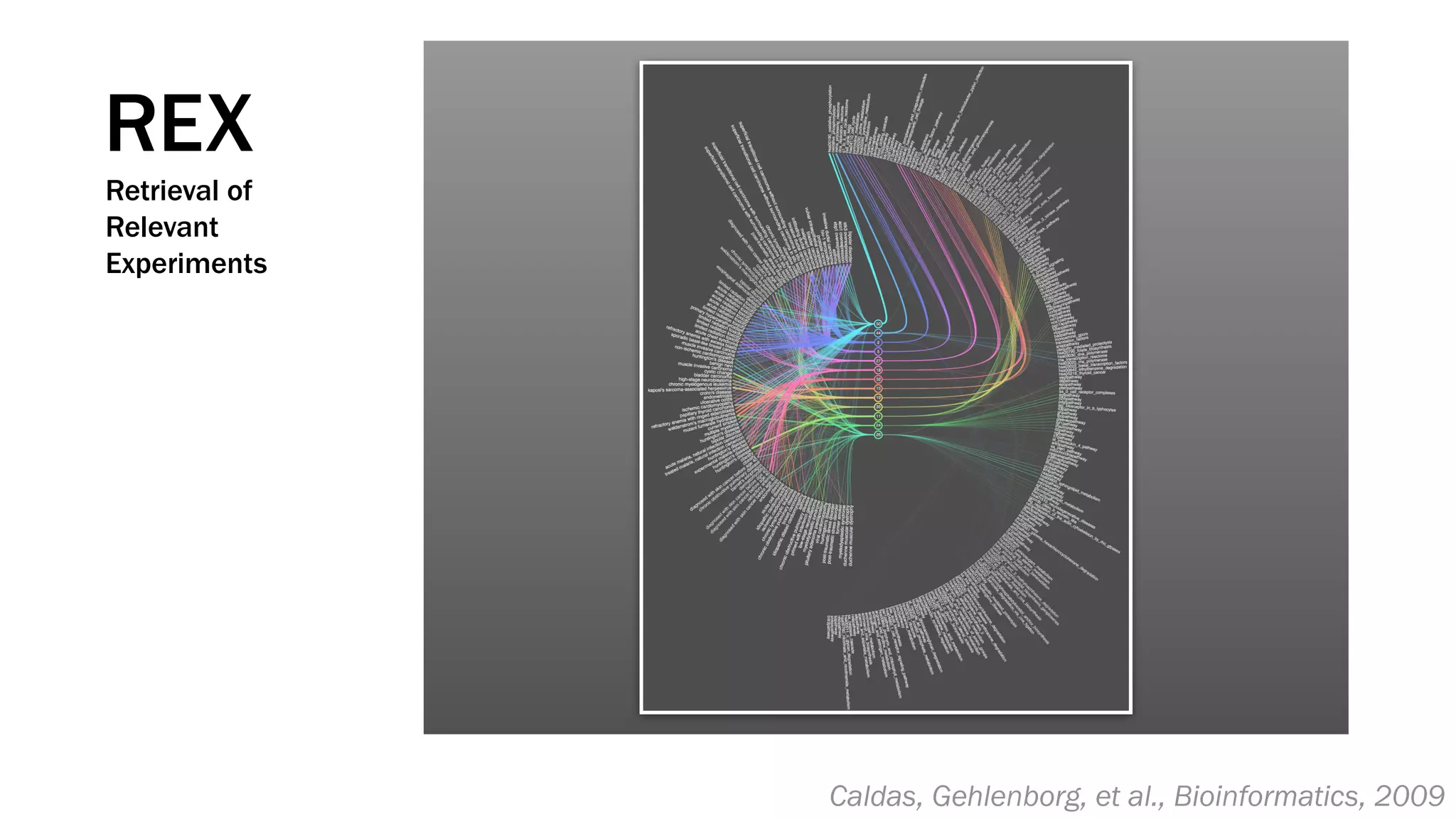
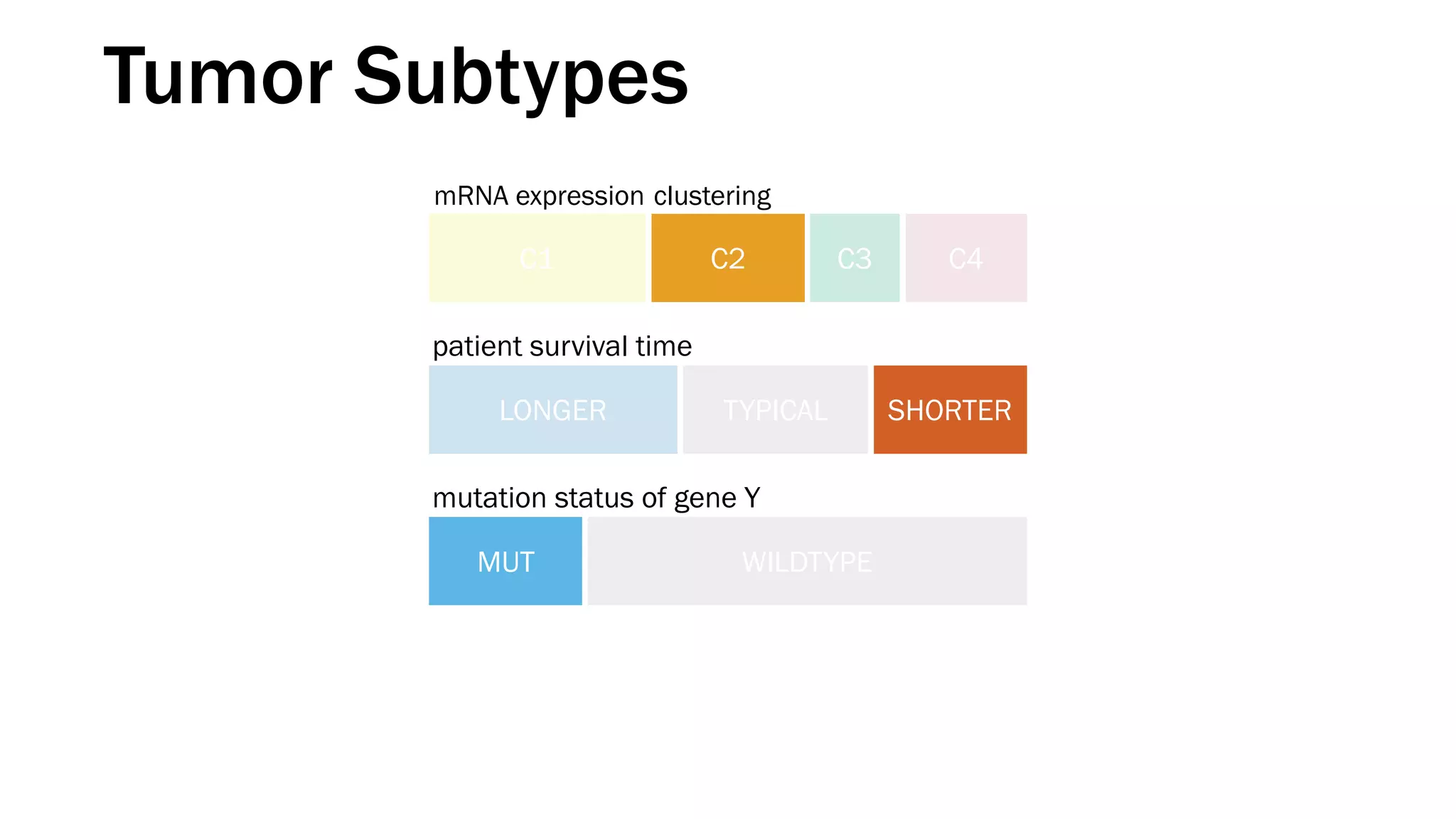
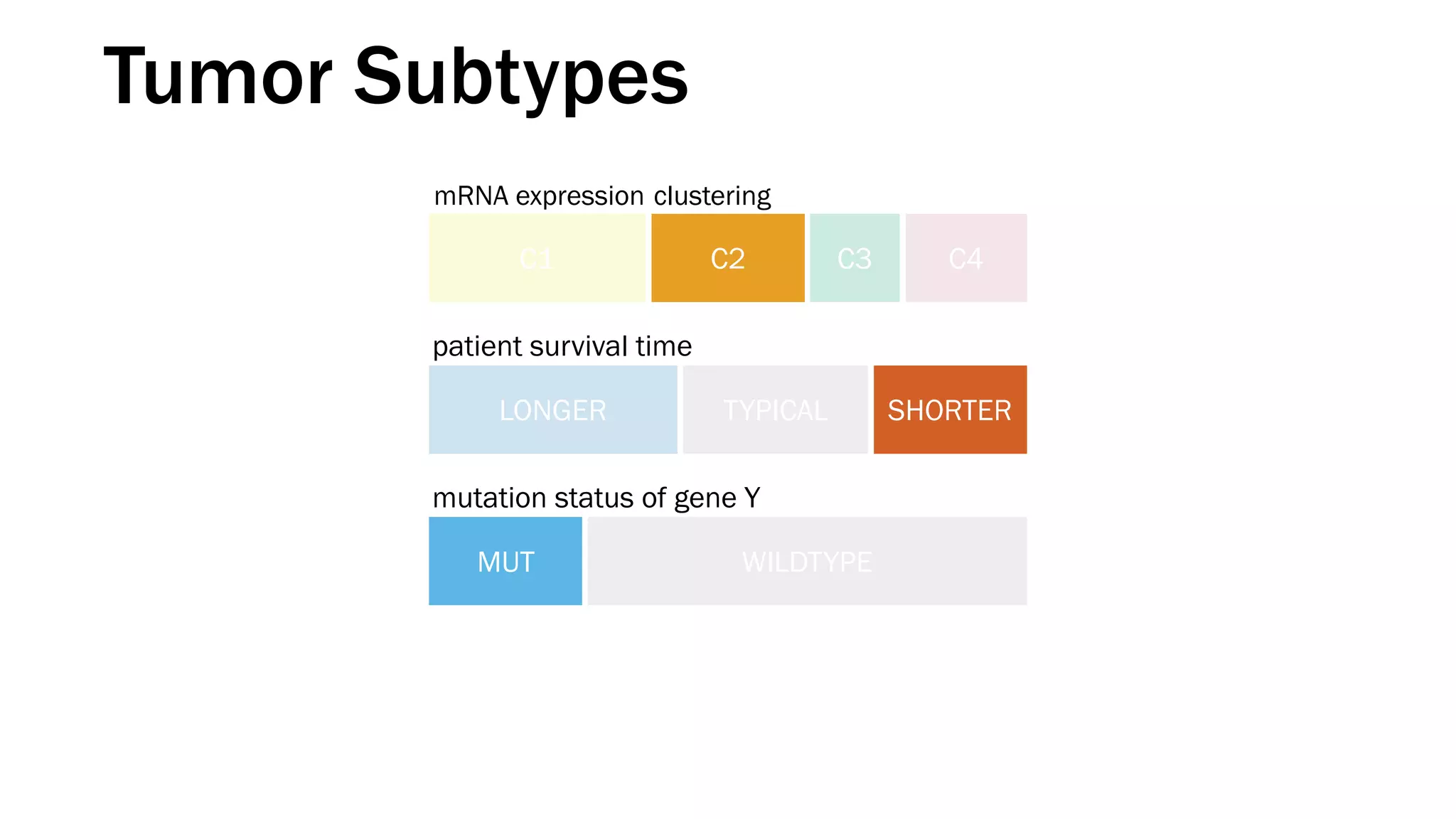
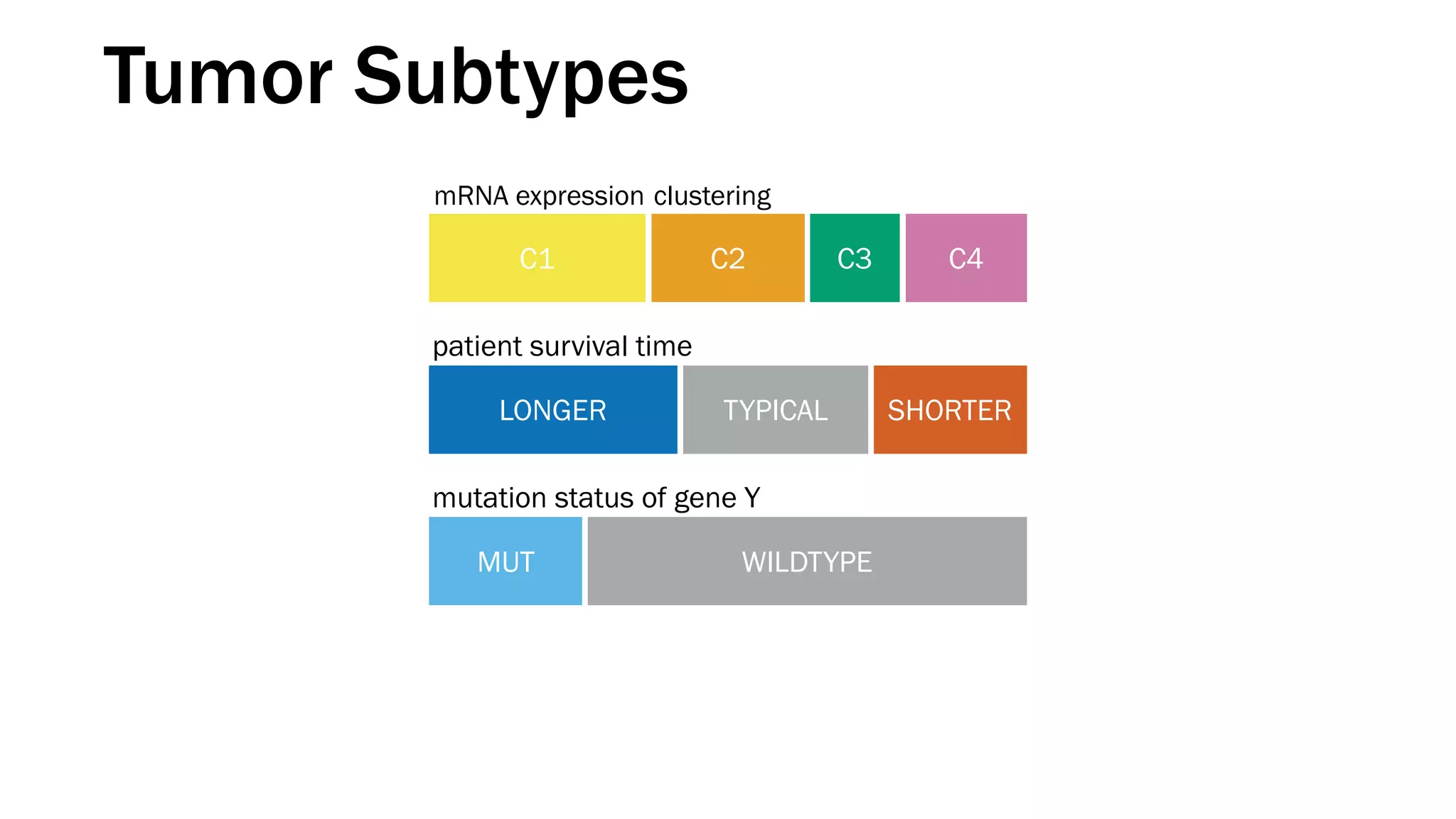
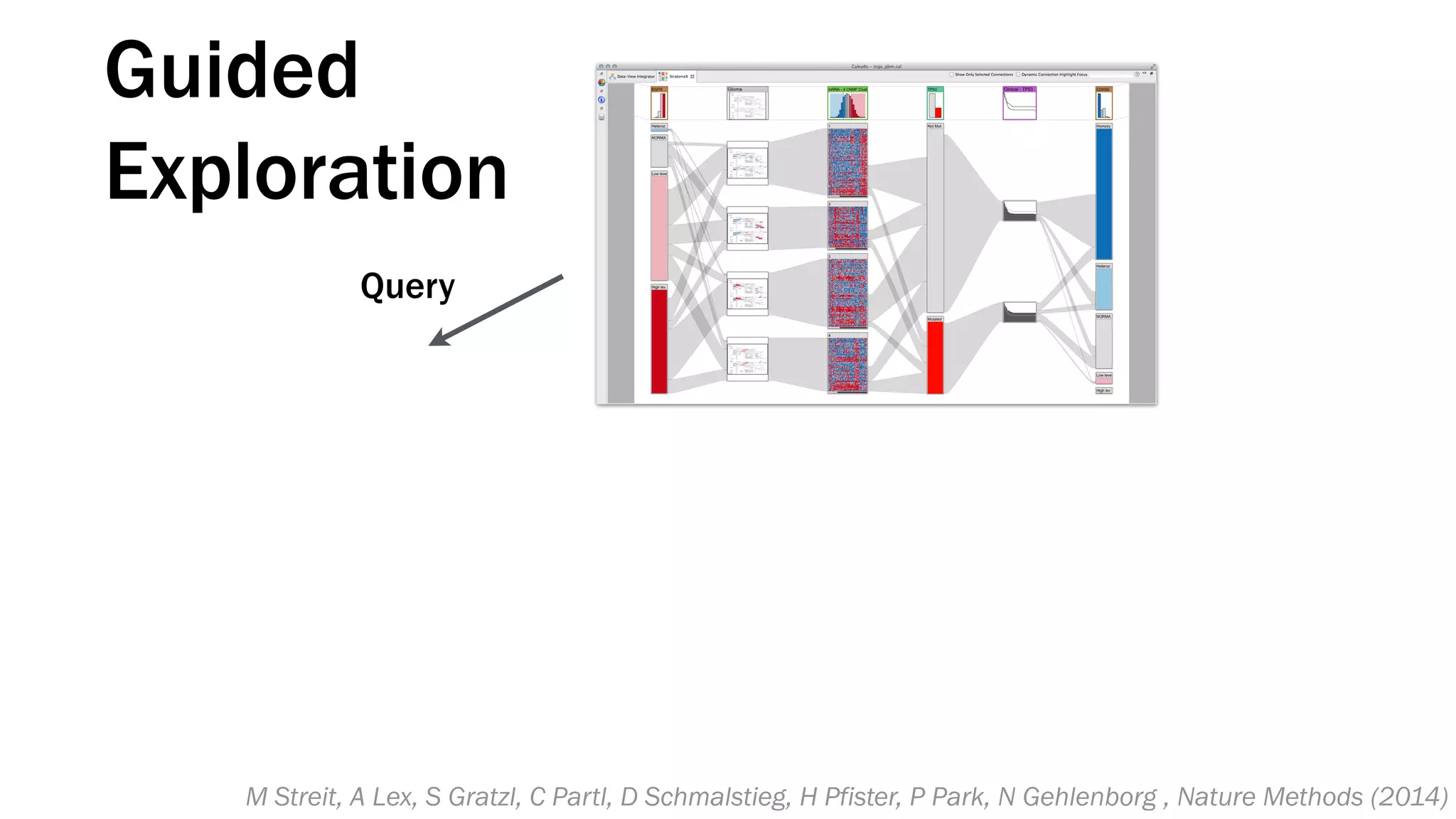
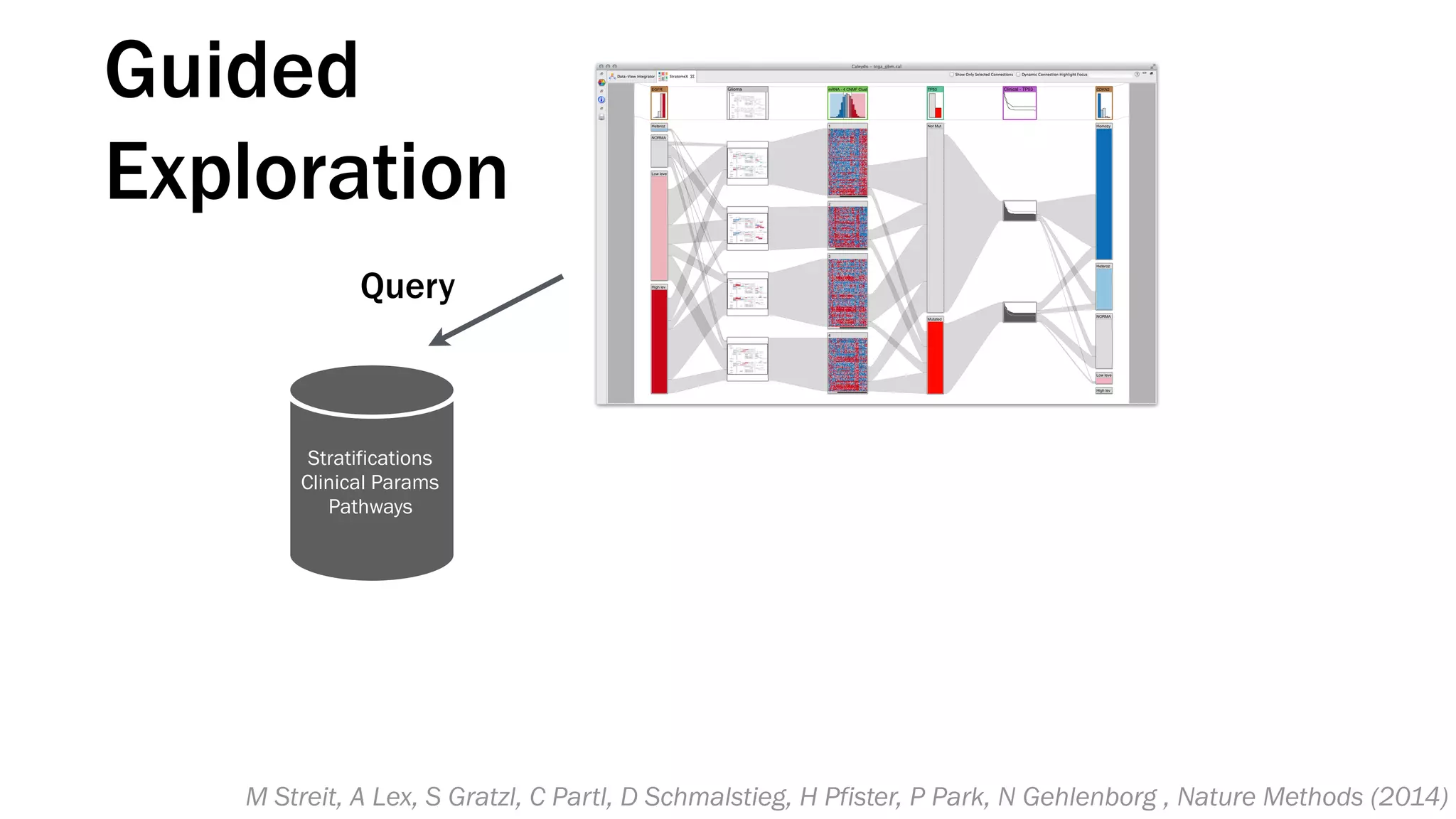
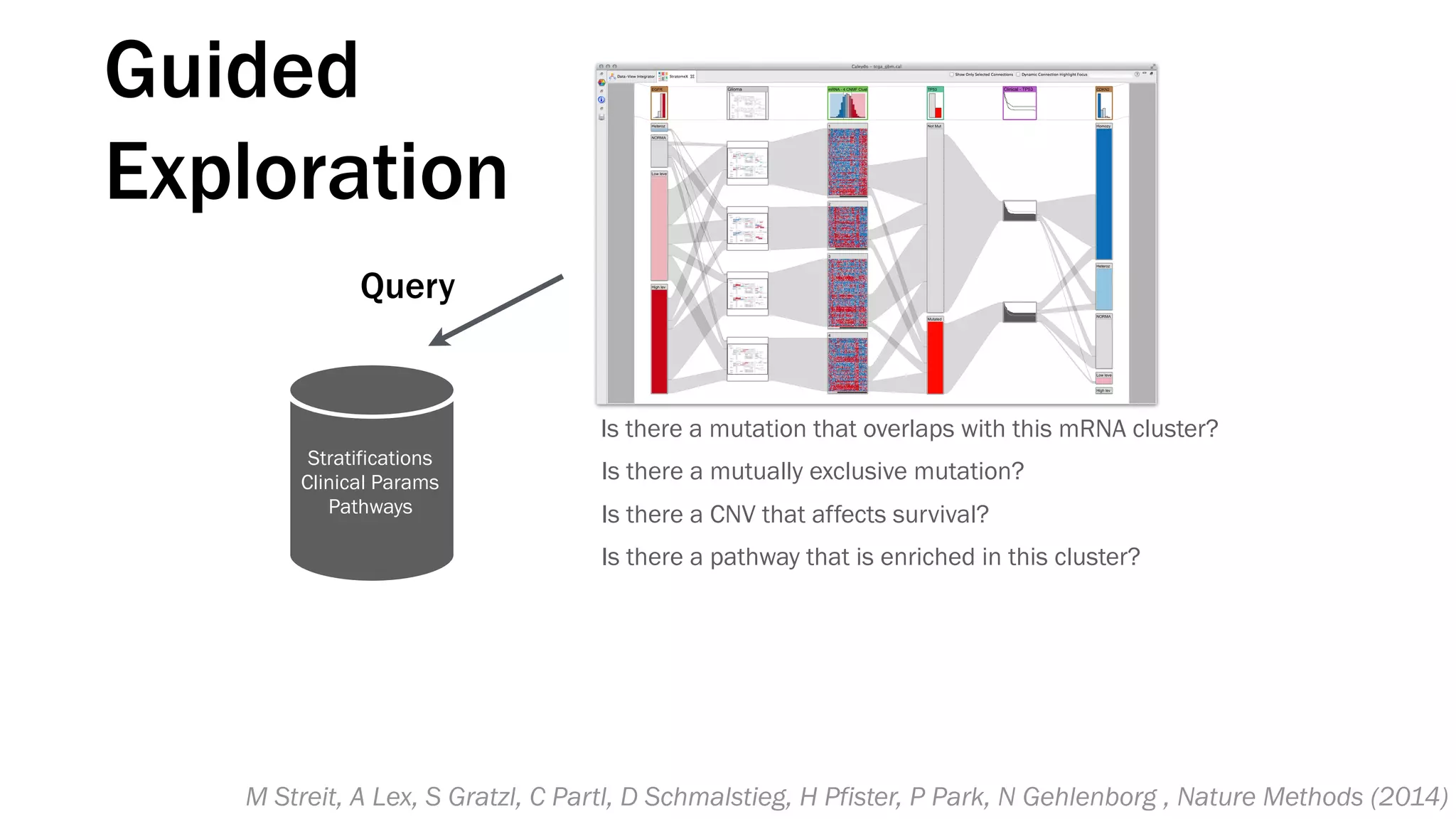
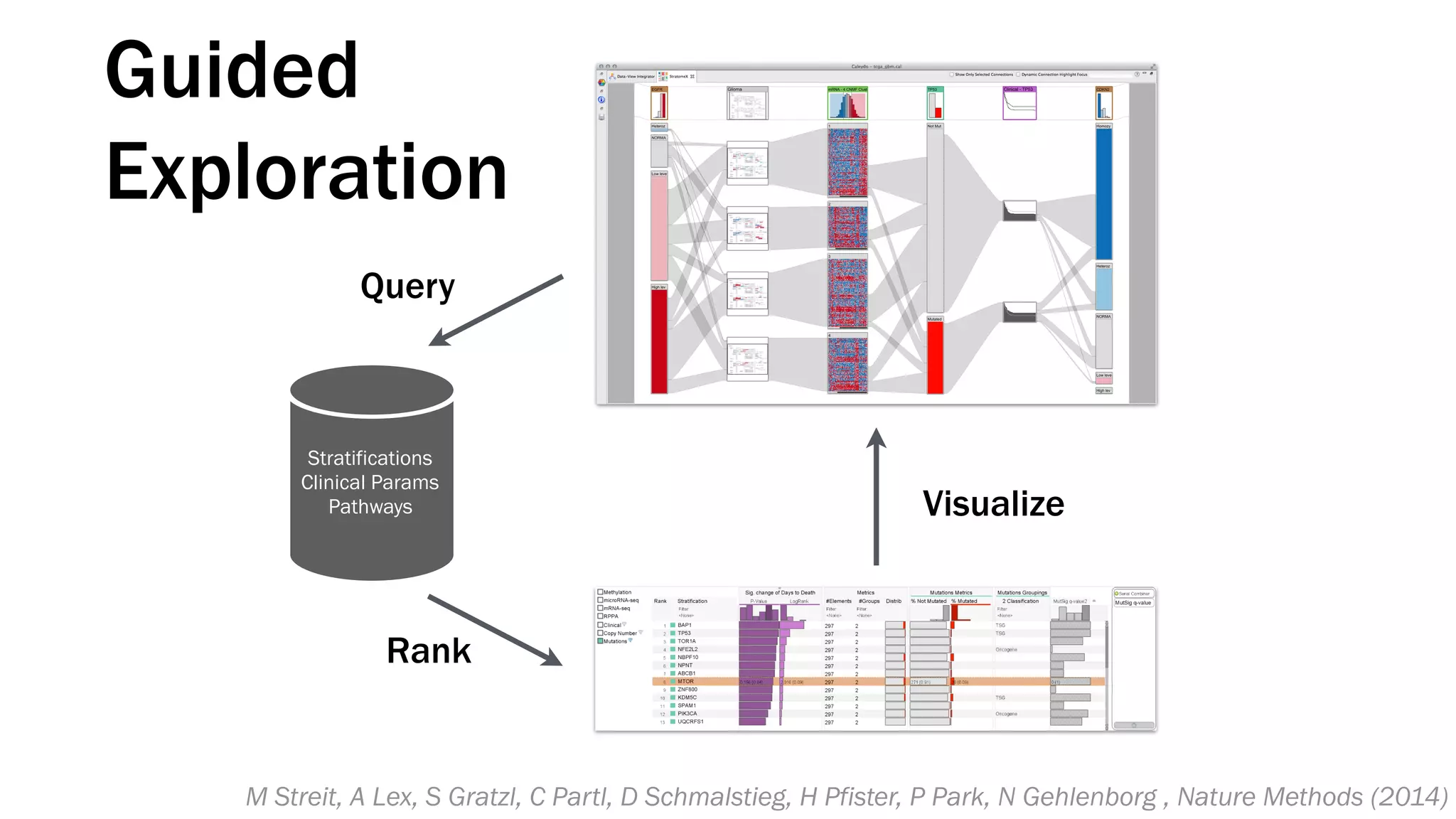
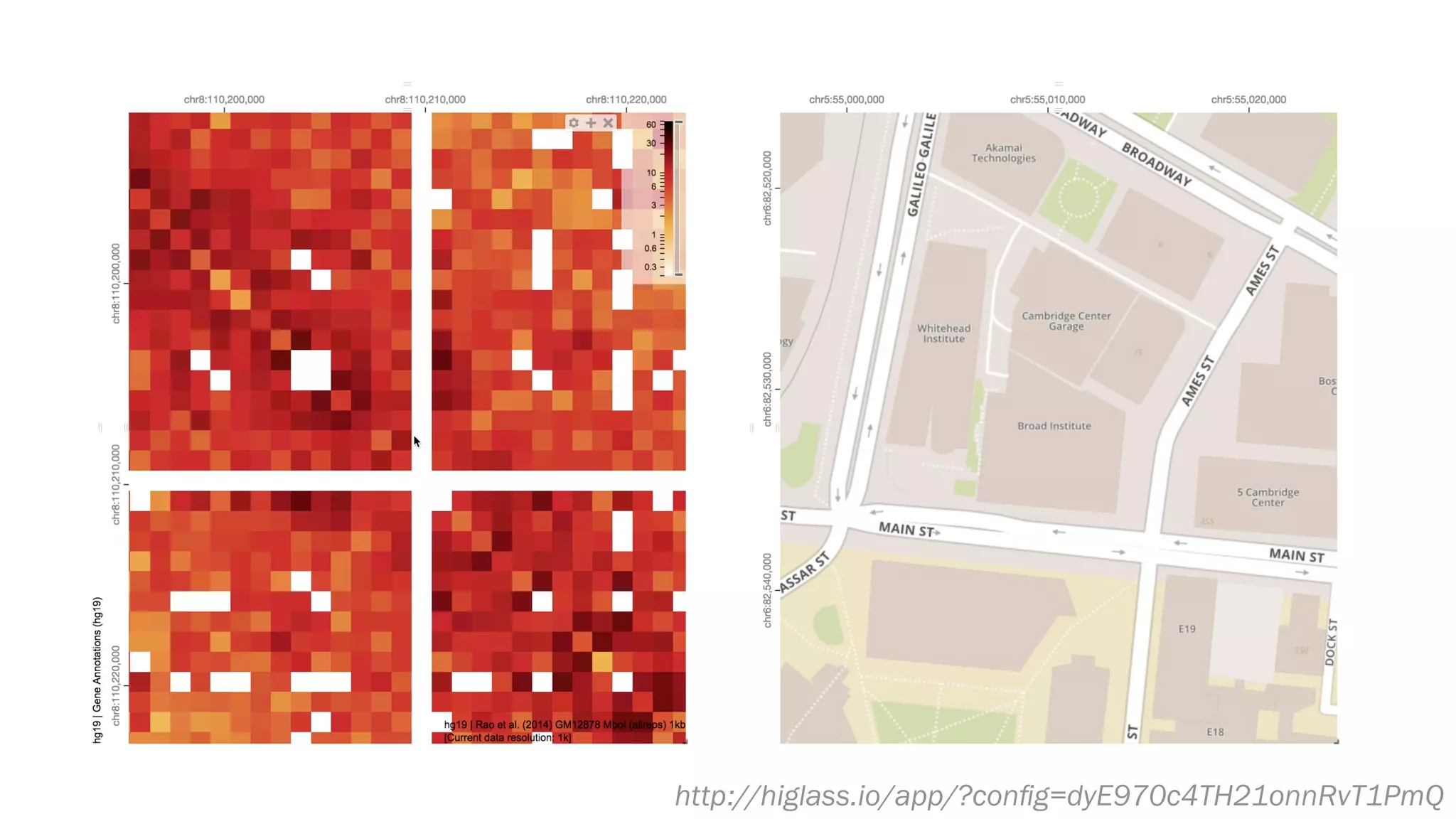
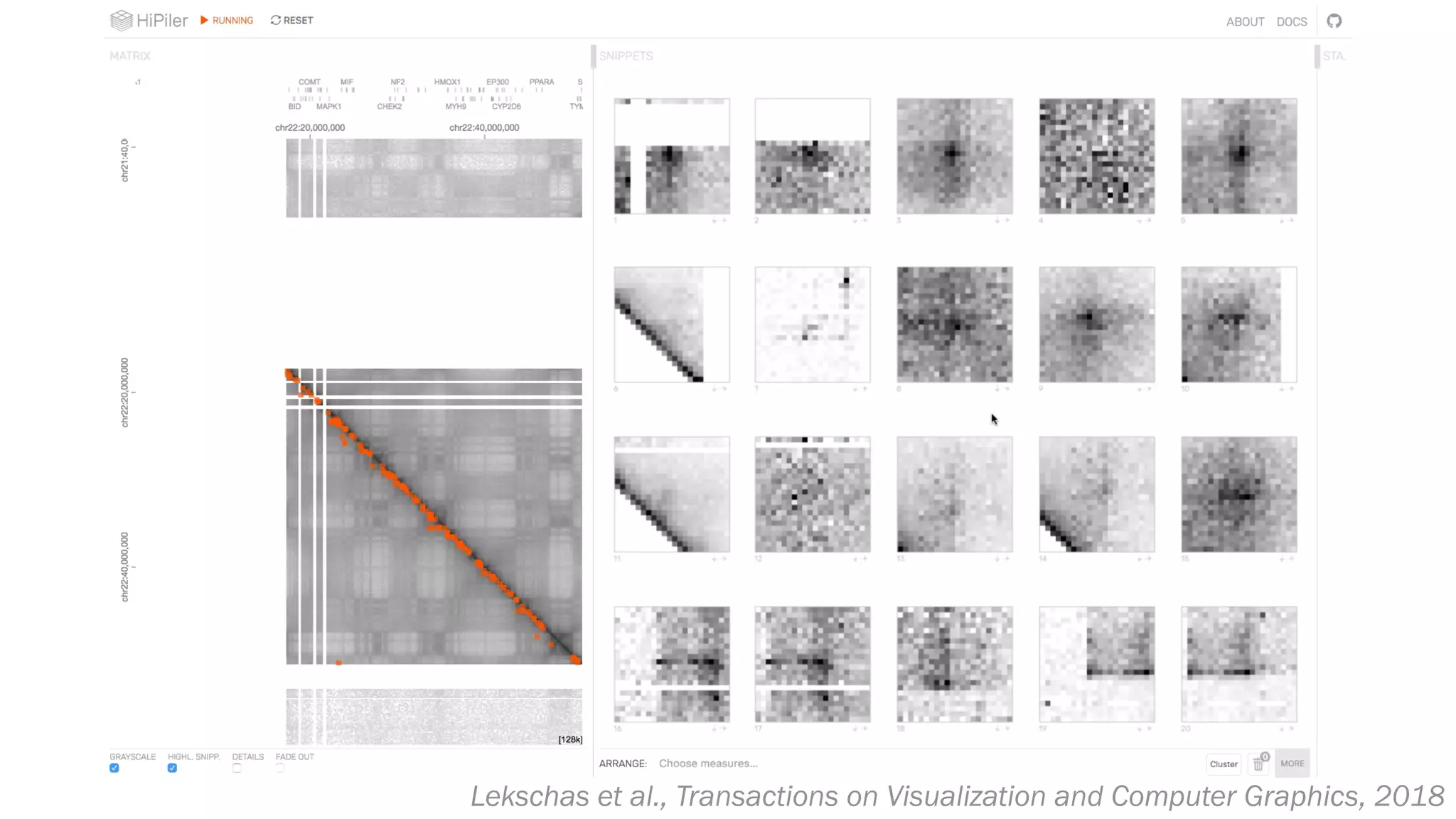
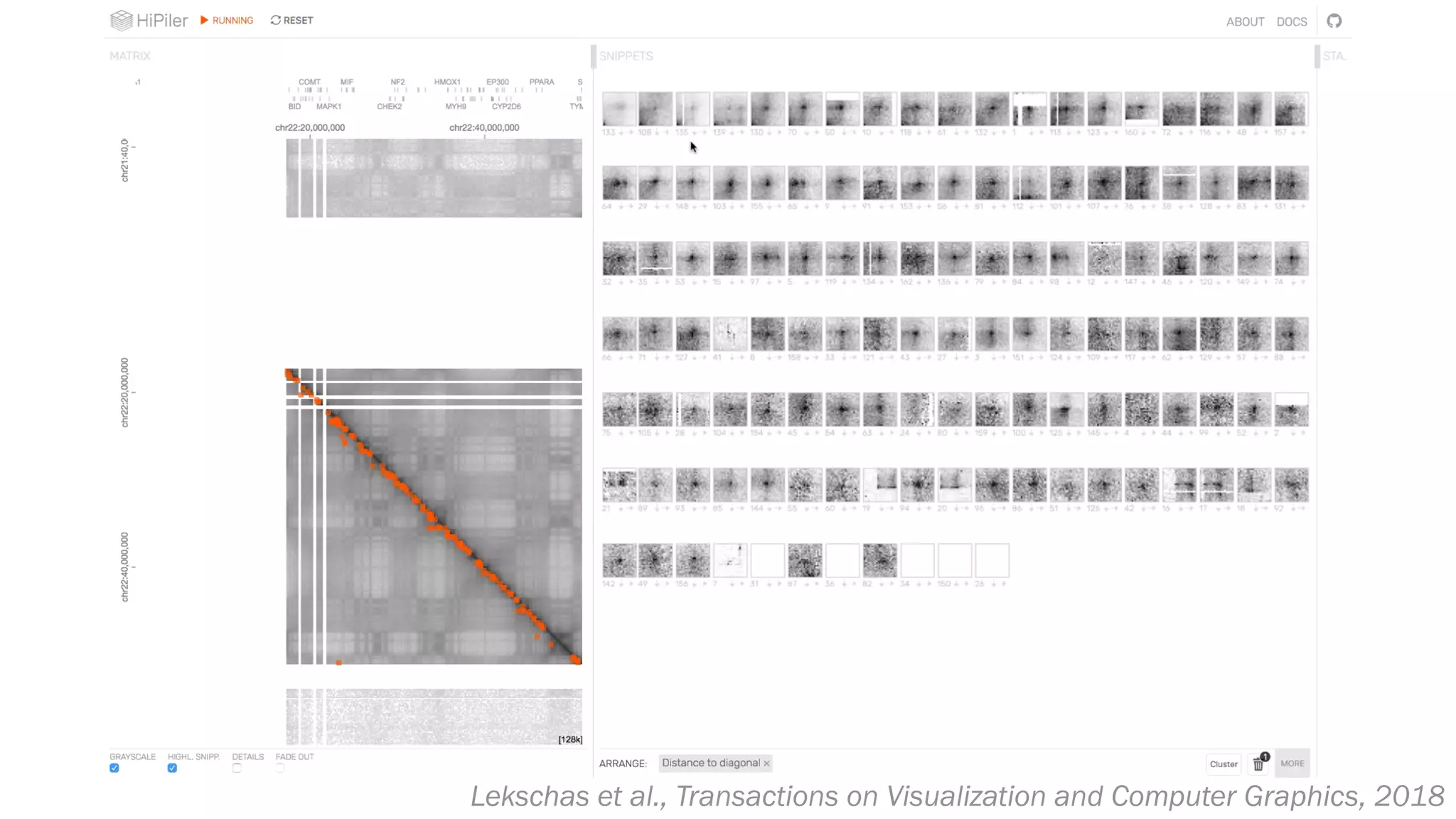
The document discusses various approaches to visualizing and interpreting transcriptomics and genomic data, highlighting the importance of hypothesis-driven and data-driven discovery. It emphasizes tools and methods developed by researchers, particularly in relation to The Cancer Genome Atlas and the Stratomex framework for patient stratifications. Additionally, it mentions collaborative contributions and the significance of open science in education and community building in biomedical informatics.