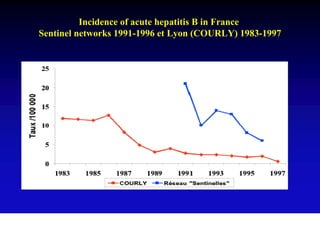
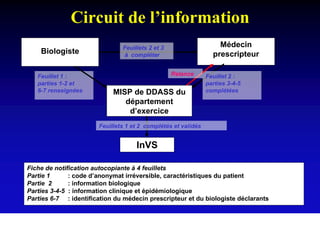
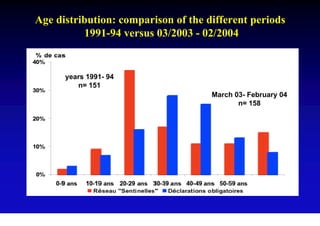
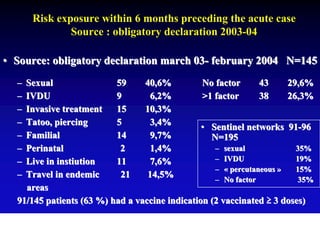
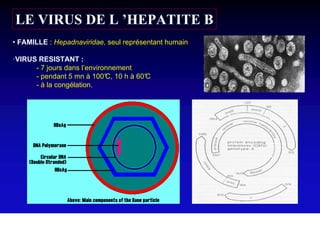
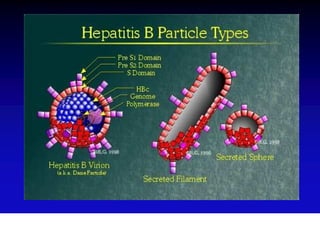
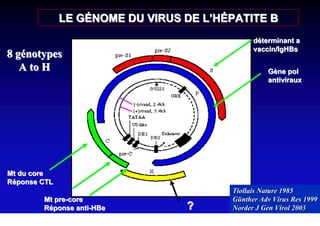
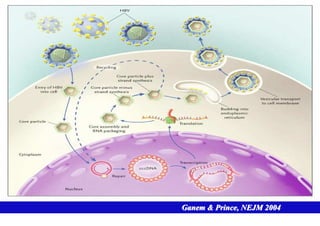
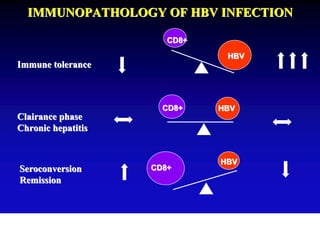
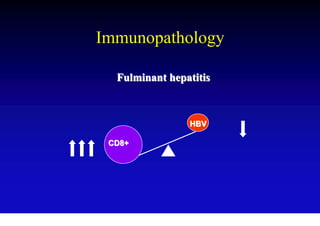
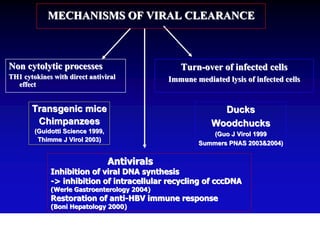
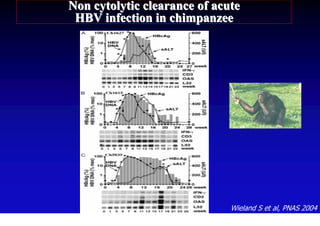
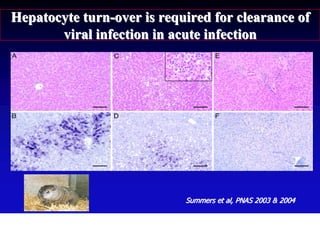
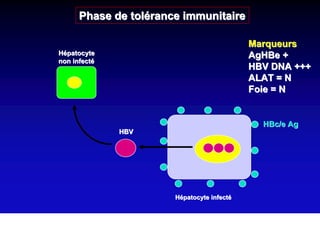
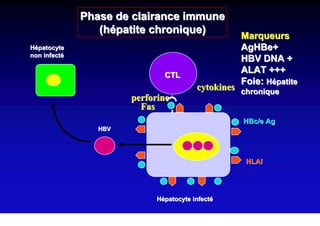
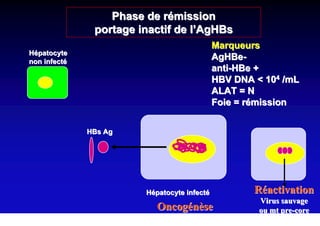
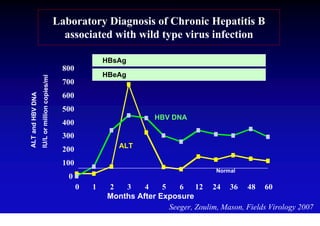
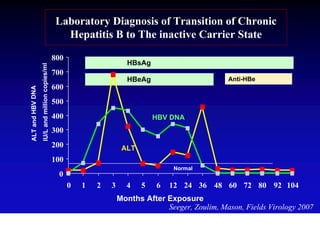
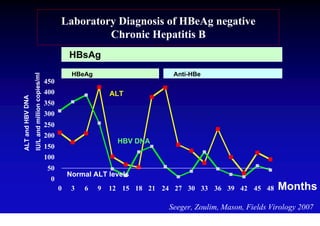
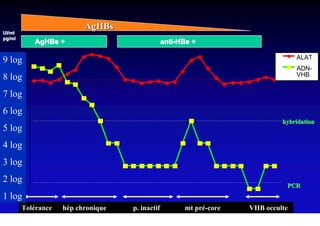
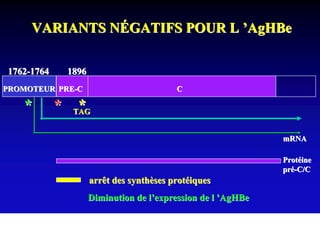
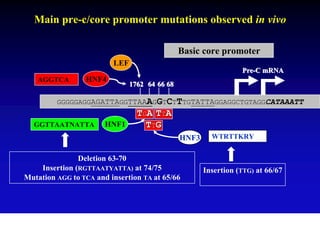
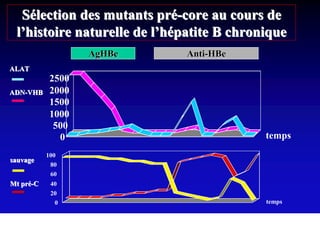
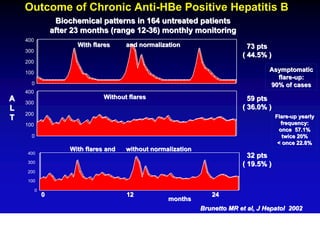
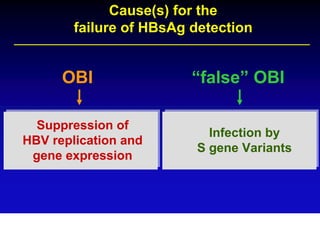
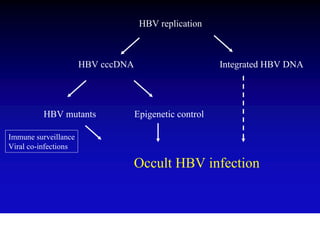
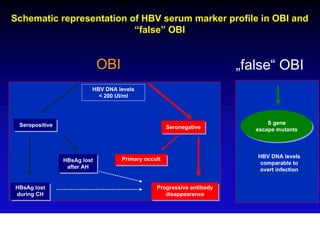
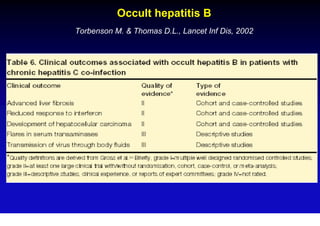
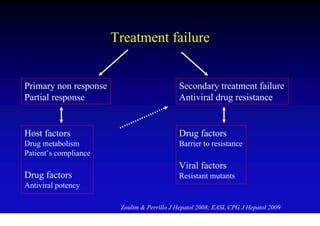
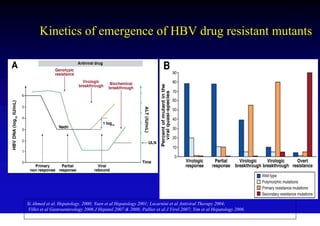
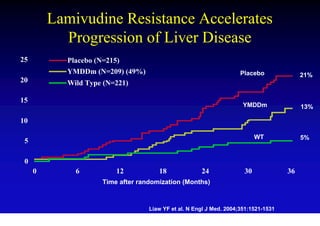
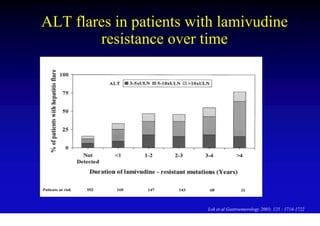
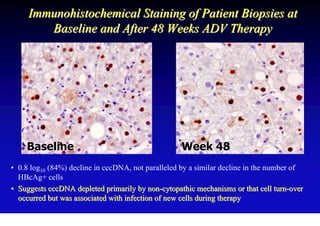
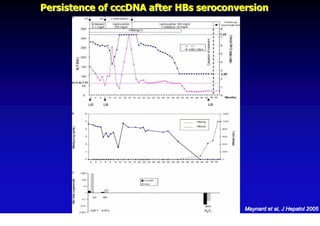
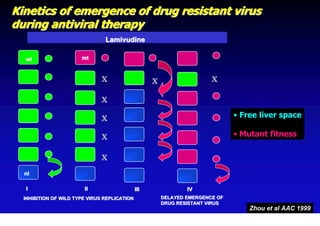
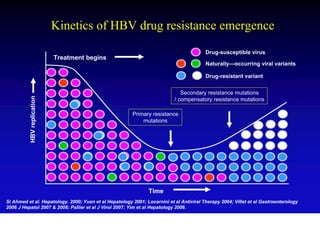
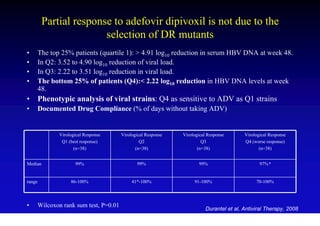
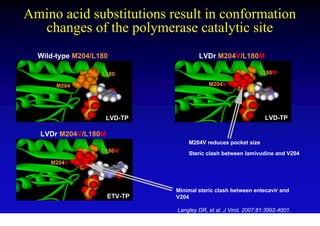
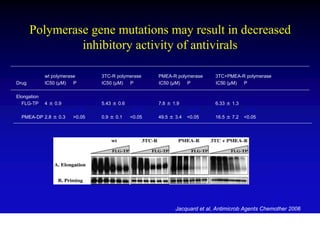
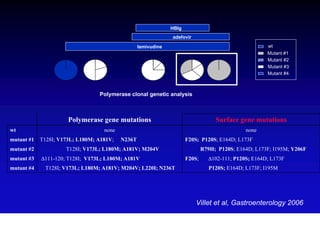
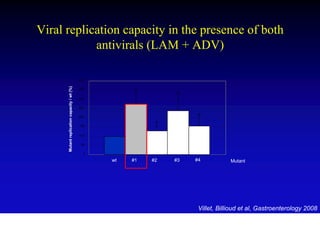
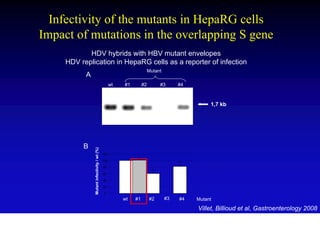
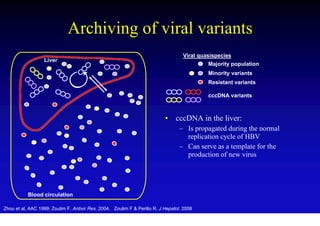
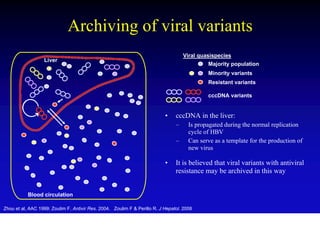
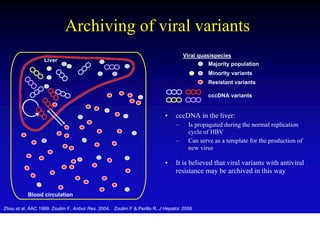
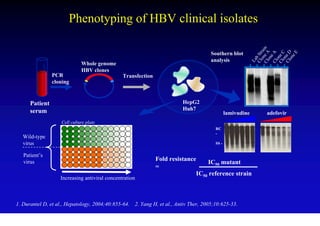
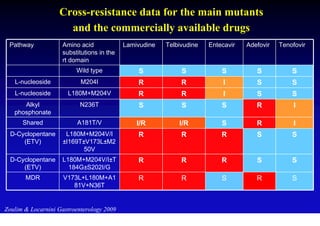
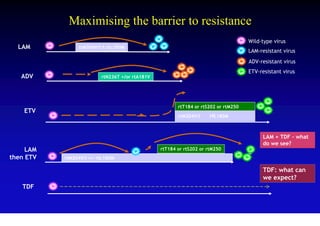
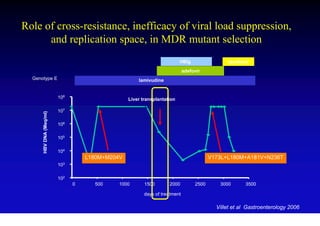
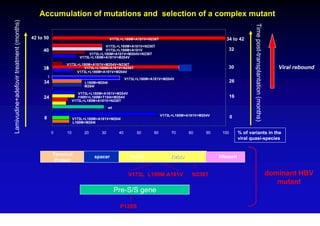
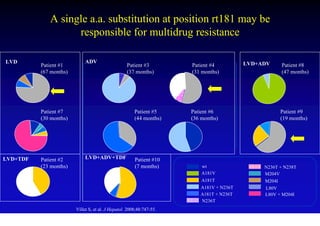
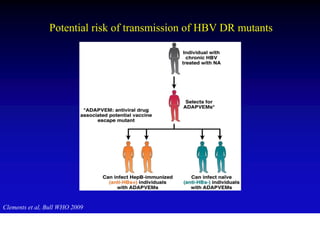
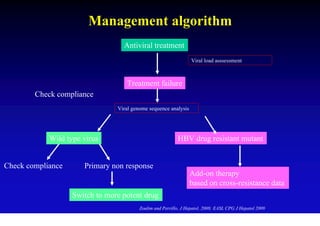
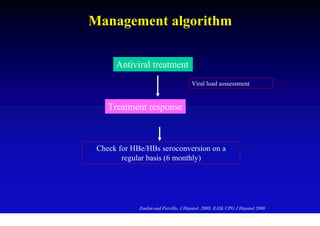
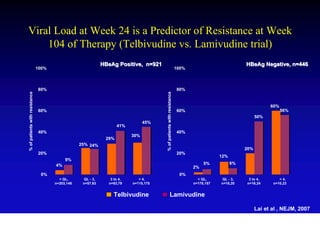
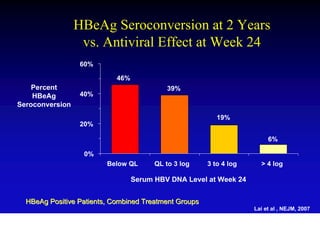
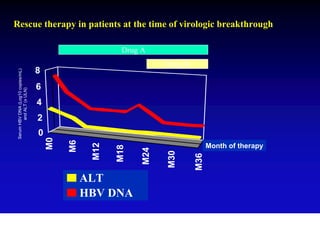
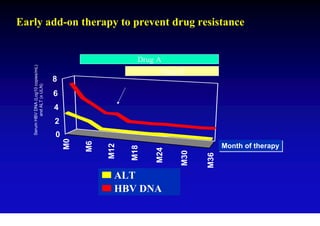
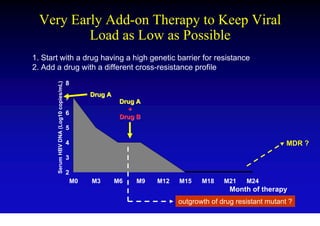
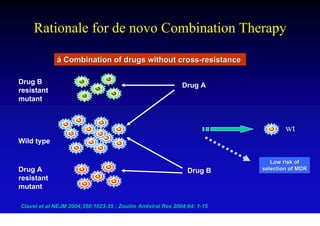
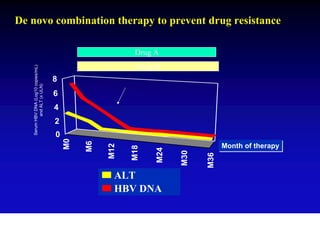
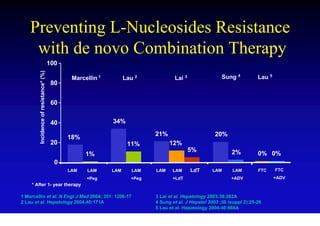
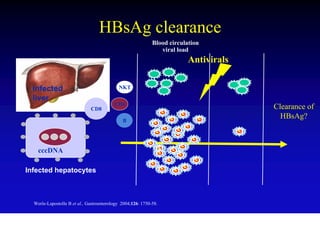
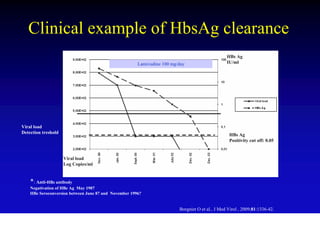
This document discusses hepatitis B, including its transmission, epidemiology, virology, and relationship to liver cancer. Some key points covered are:
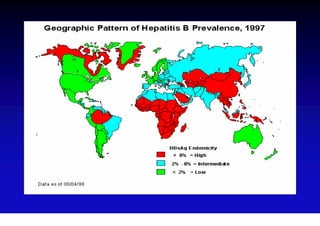
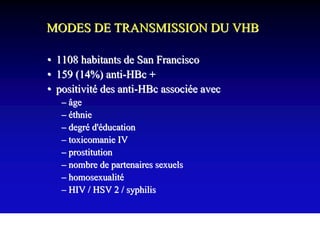
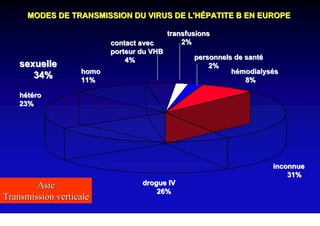
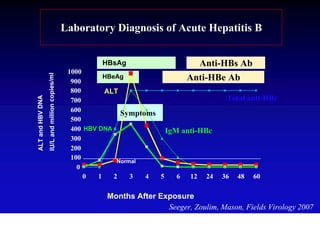
- Hepatitis B virus can be transmitted sexually, through blood exposure or mother-to-child transmission.
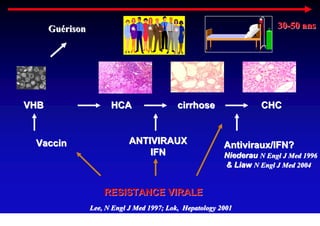
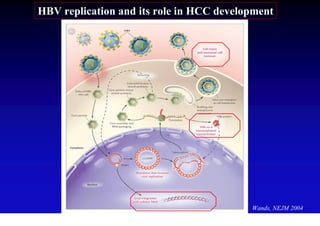
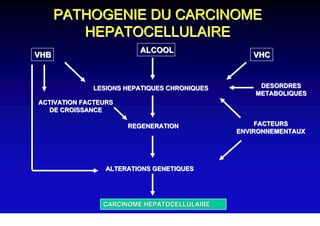
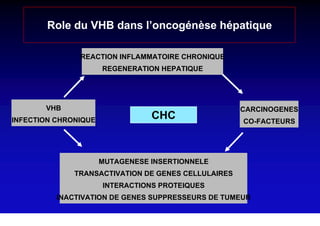
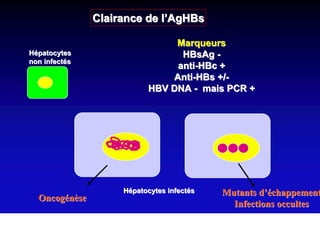
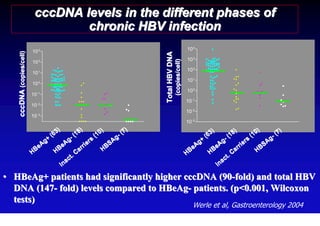
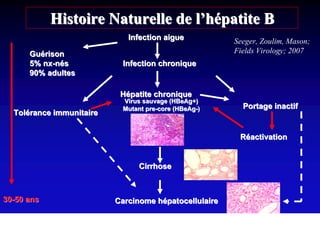
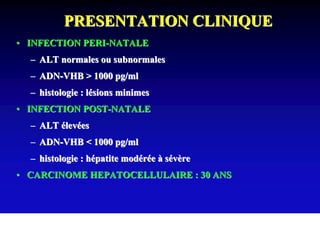
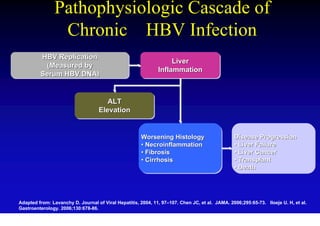
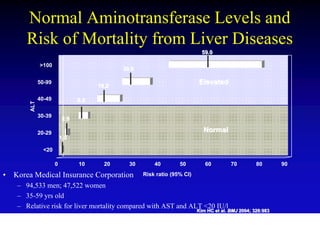
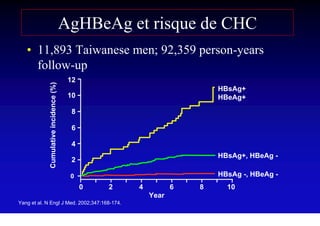
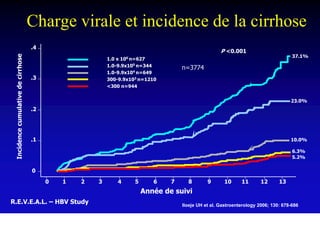
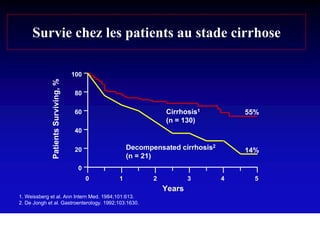
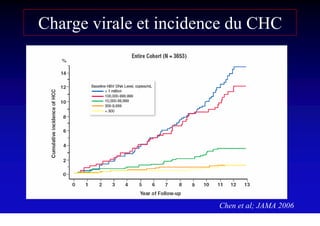
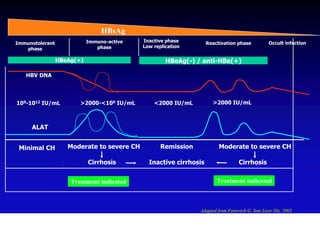
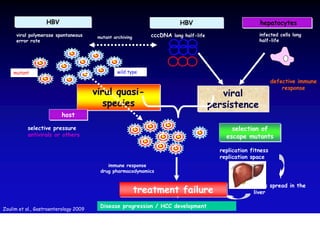
- Chronic hepatitis B infection can lead to cirrhosis and hepatocellular carcinoma over 30-50 years.
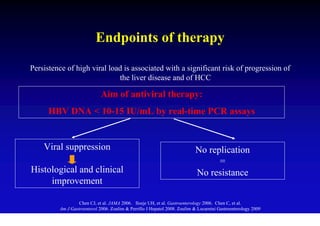
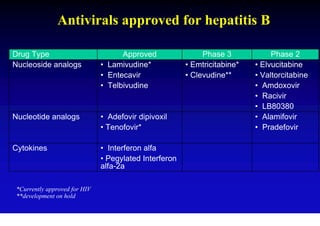
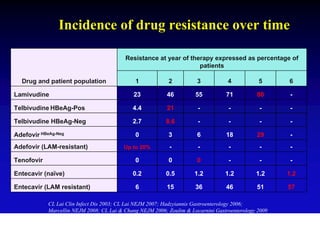
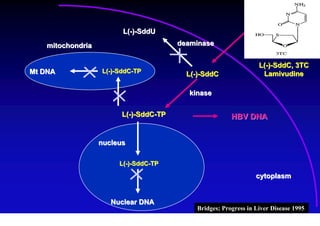
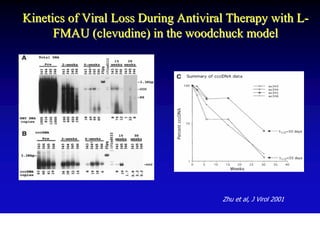
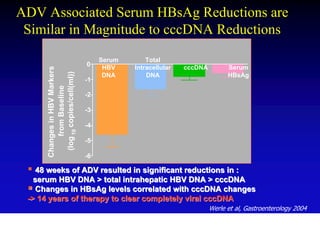
- Antiviral treatments and vaccines have reduced transmission and complications of hepatitis B.
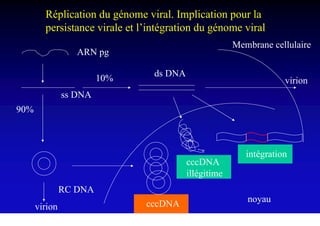
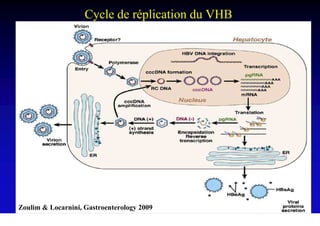
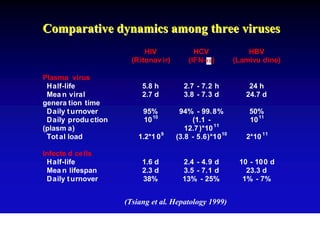
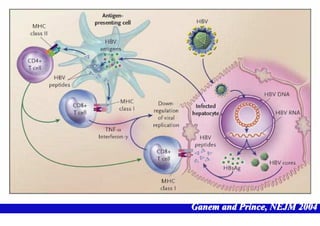
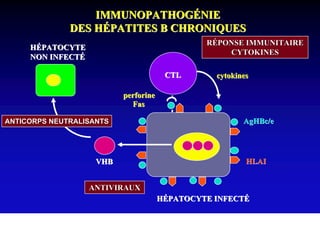
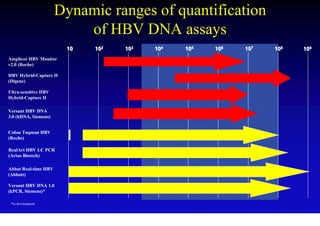
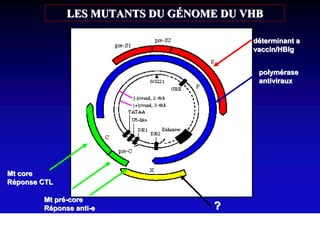
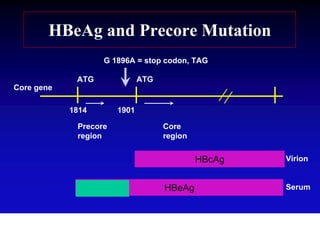
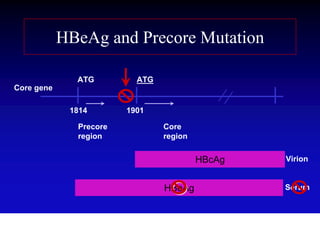
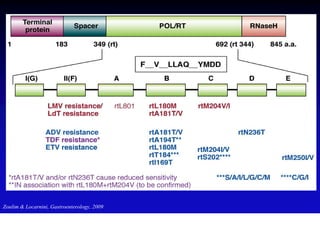
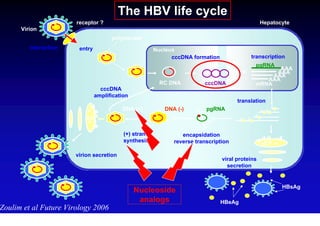
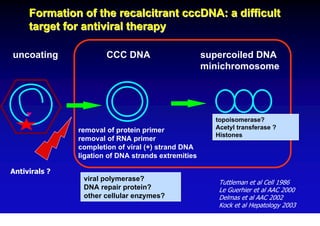
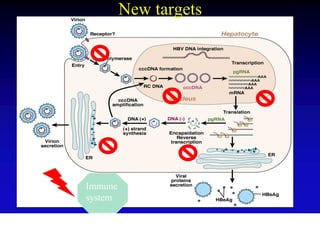
- The hepatitis B virus genome and life cycle allow it to persist chronically in the liver more than other viruses like hepatitis C and HIV.
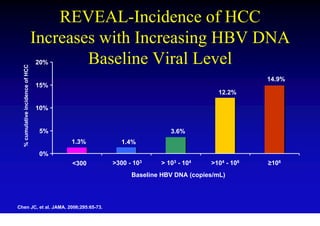
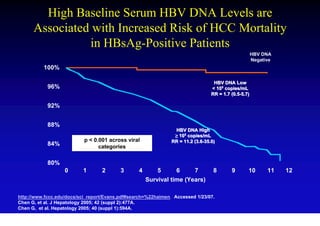
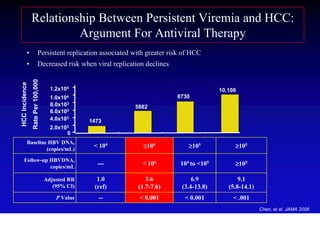
- Chronic hepatitis B is a major risk factor for the development of hepatocellular carcinoma