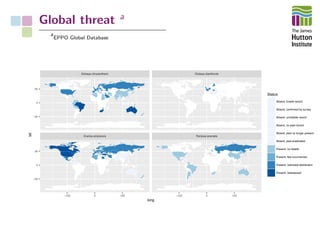
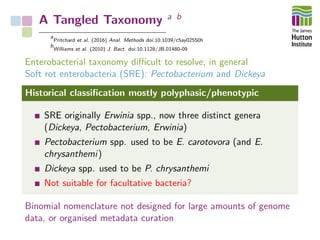
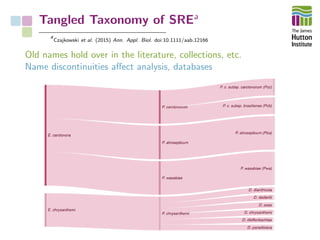
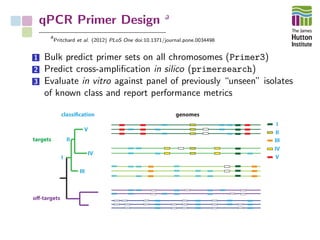
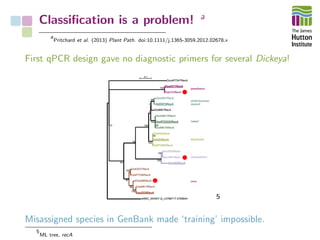
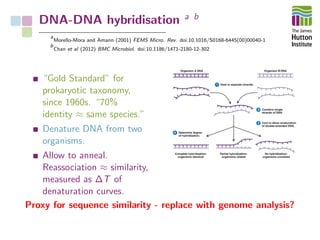
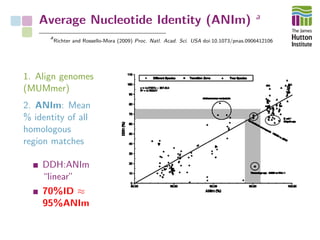
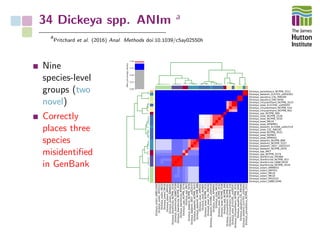
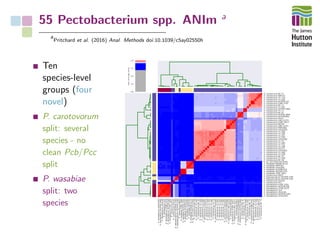
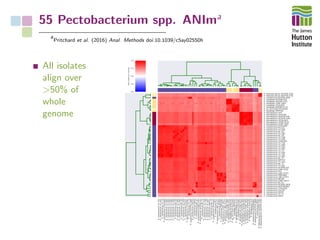
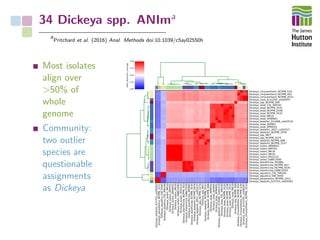
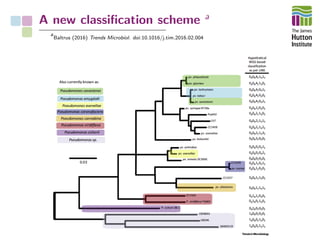
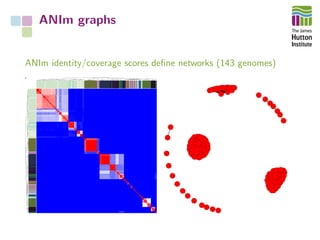
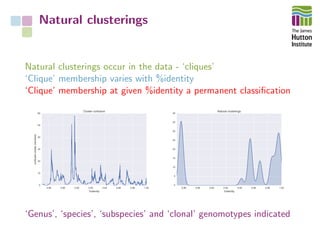
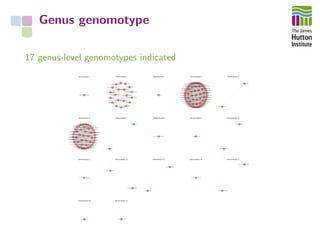
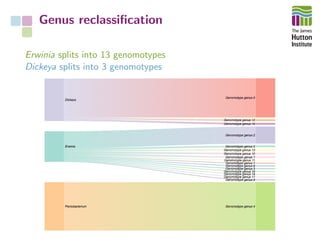
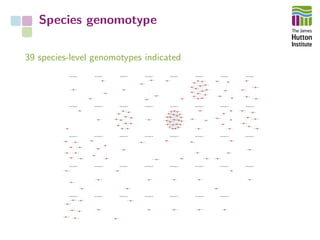
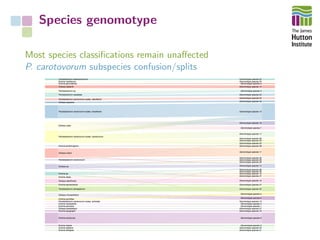
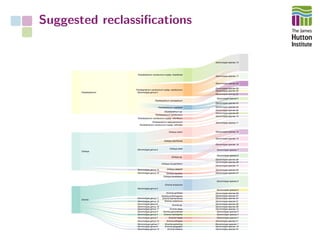
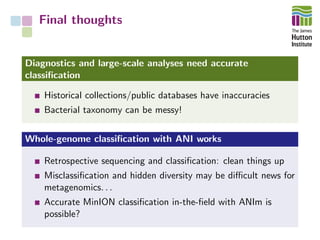
The document addresses the challenges in classifying bacterial potato pathogens, particularly within the genera Dickeya and Pectobacterium, revealing a tangled taxonomy rooted in historical names and classifications. It discusses the implications of accurate classification for quarantine and disease management, emphasizing the need for effective diagnostic tools like qPCR. The presentation highlights recent advancements in whole genome analysis and the application of Average Nucleotide Identity for more precise pathogen identification.