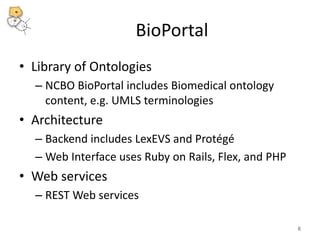
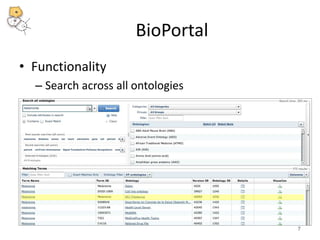
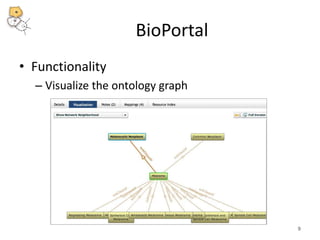
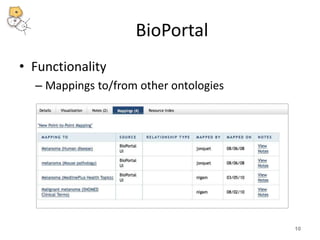
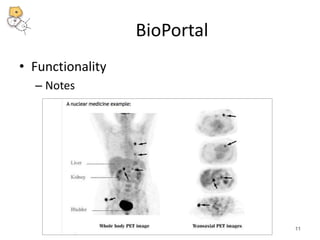
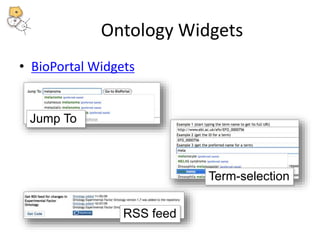
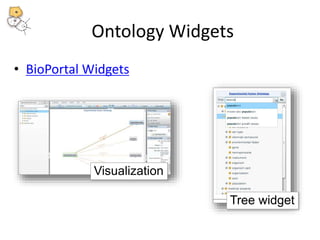
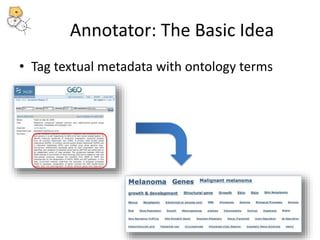
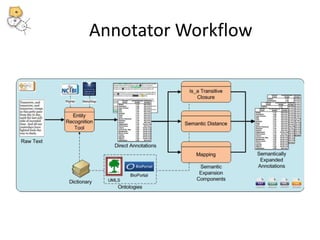
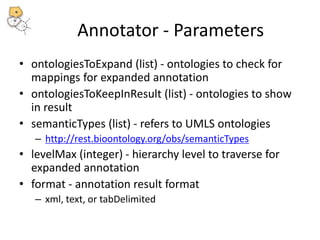
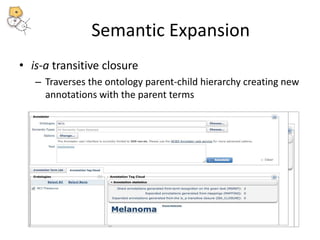
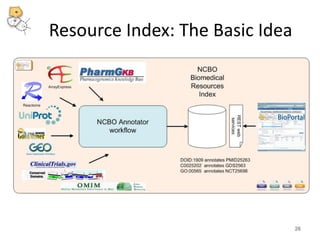
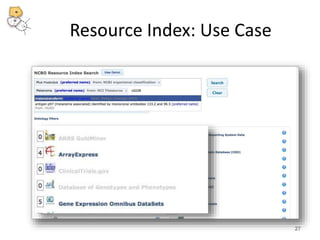
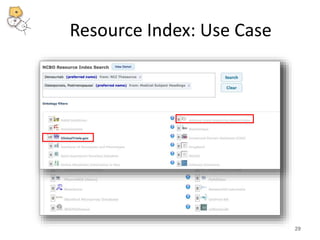
The National Center for Biomedical Ontology (NCBO) provides several software tools to enable semantically aware applications. These include BioPortal, an online library of biomedical ontologies, Ontology Widgets which allow integration of ontologies into websites, and the Annotator, a web service that semantically tags text with ontology terms. The NCBO also plans to develop a comprehensive index of online biomedical resources.