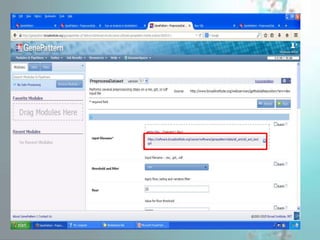
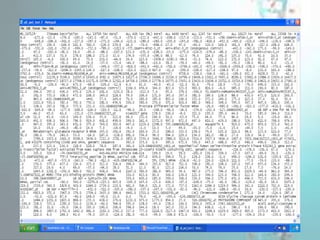
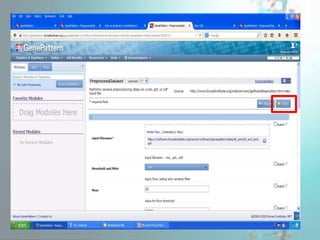
GenePattern is a freely available software package released in 2004 that provides access to over 250 genomic analysis tools. It allows users to design sophisticated analysis pipelines by chaining modules together to capture methods, parameters, and data used to produce results. Pipelines can be used to create, edit, share, and reproduce in silico genomic analysis results.