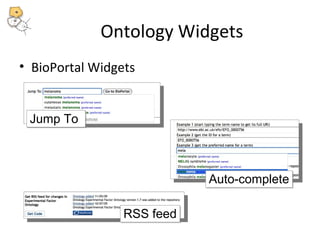
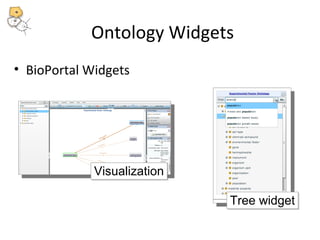
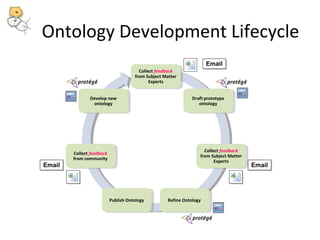
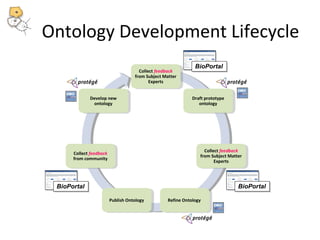
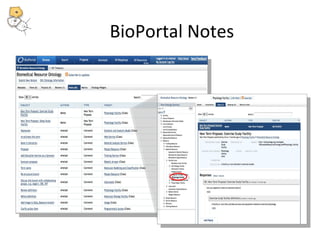
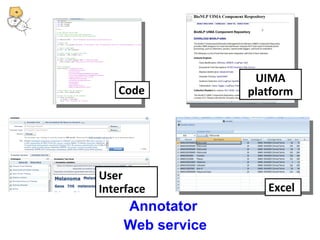
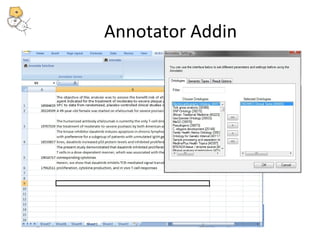
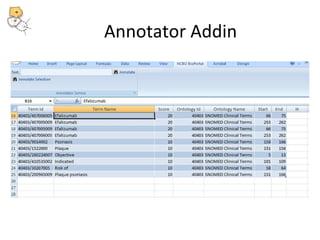
This document discusses tools from the National Center for Biomedical Ontology (NCBO) that can enhance the data curation workflow. It describes Ontology Widgets and web services that allow consistent annotation of data by reusing terms from ontologies. It also describes BioPortal Notes for collecting structured feedback to enrich ontologies, and the NCBO Annotator web service for annotating textual metadata with ontology terms. These tools address challenges of curating large amounts of data by facilitating data submission, ontology evolution, and metadata annotation.