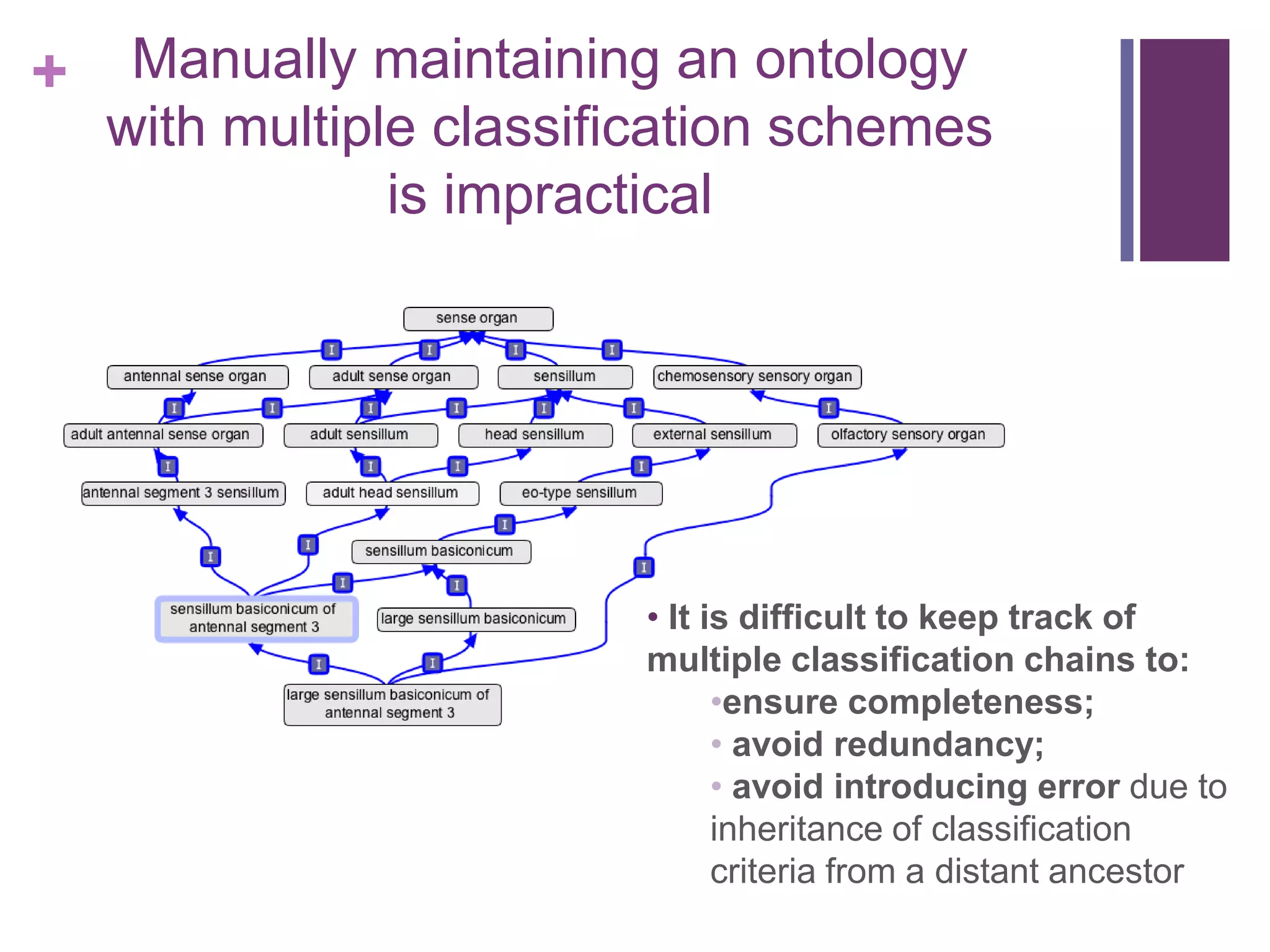
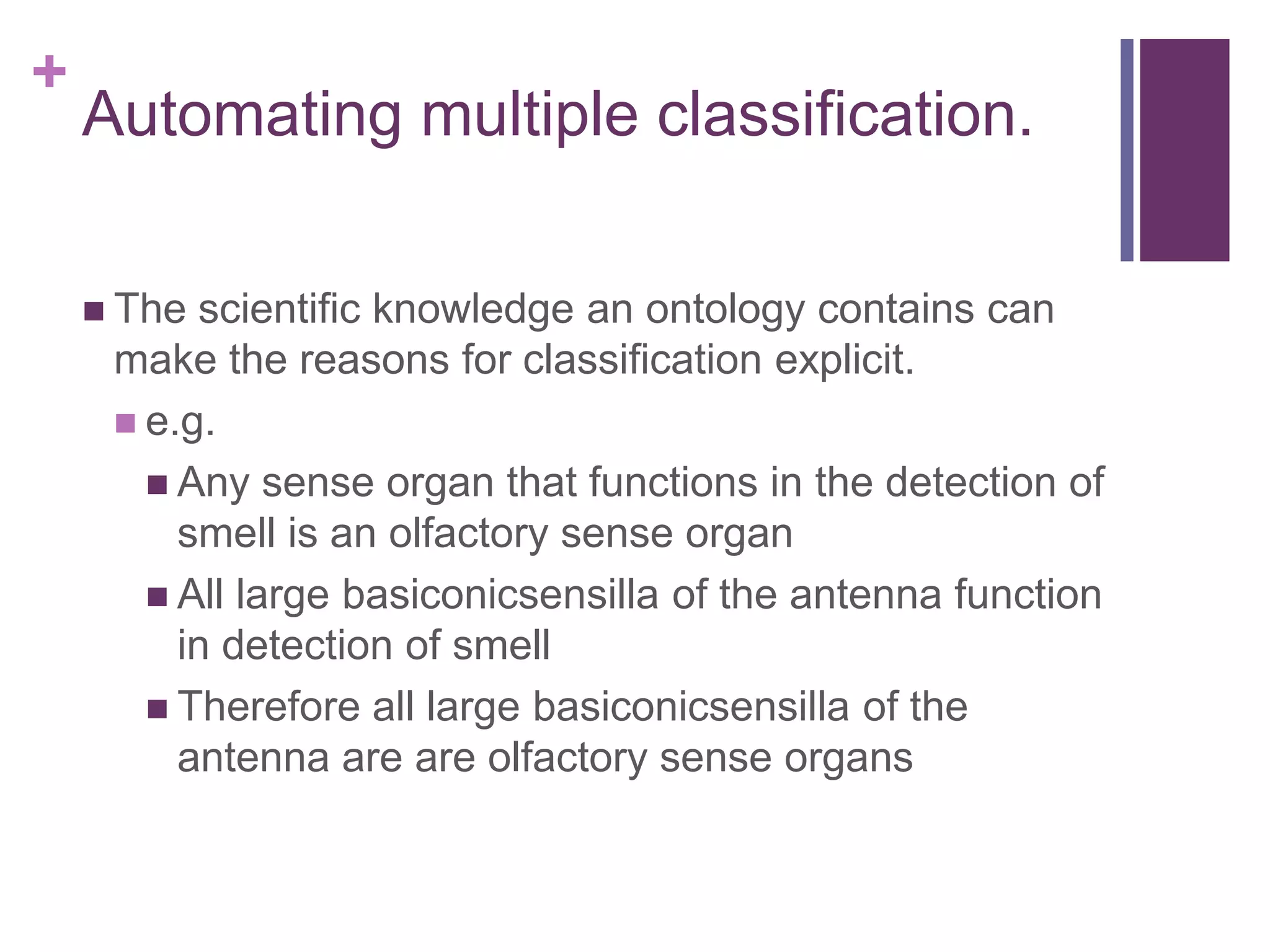
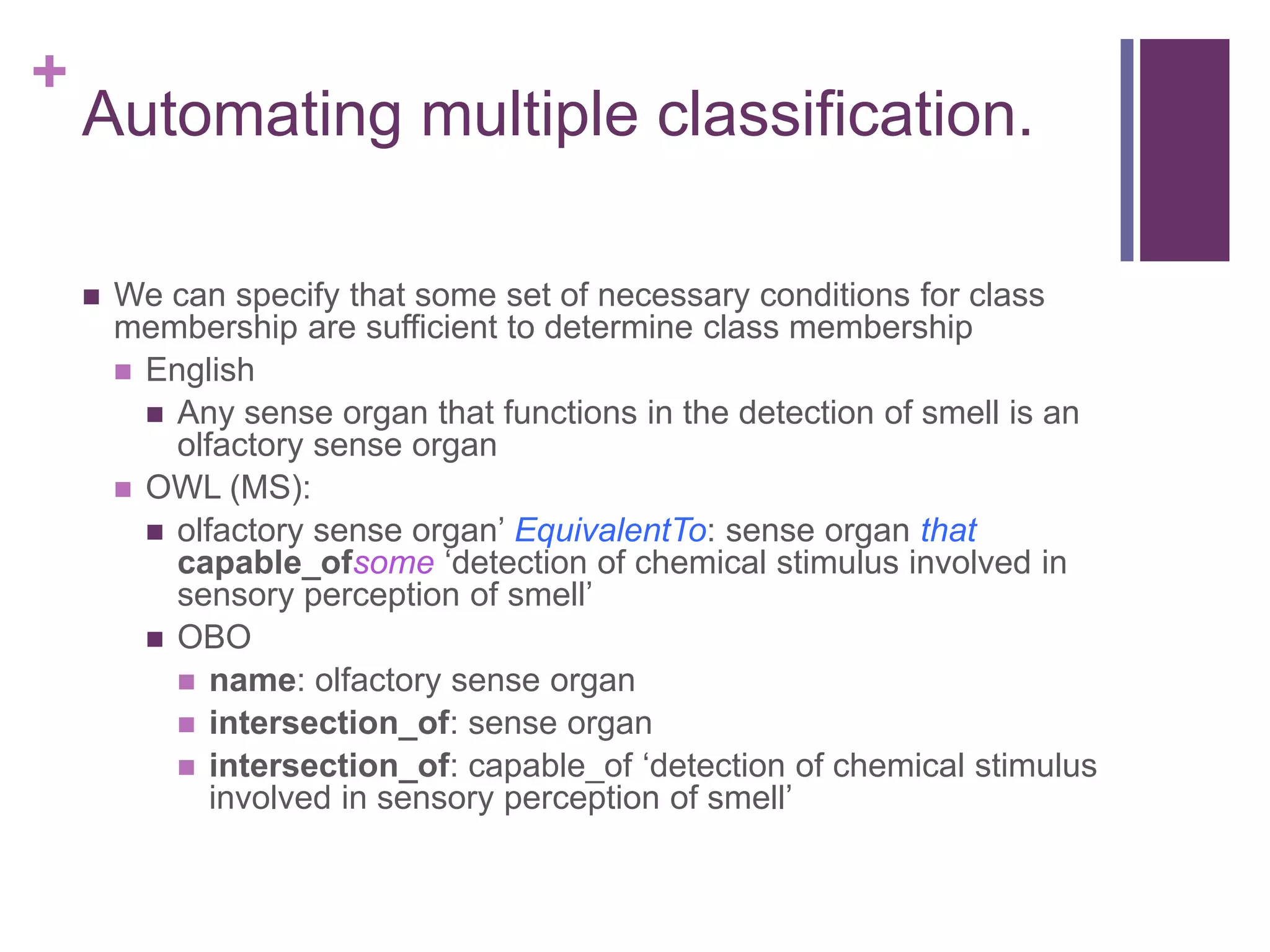
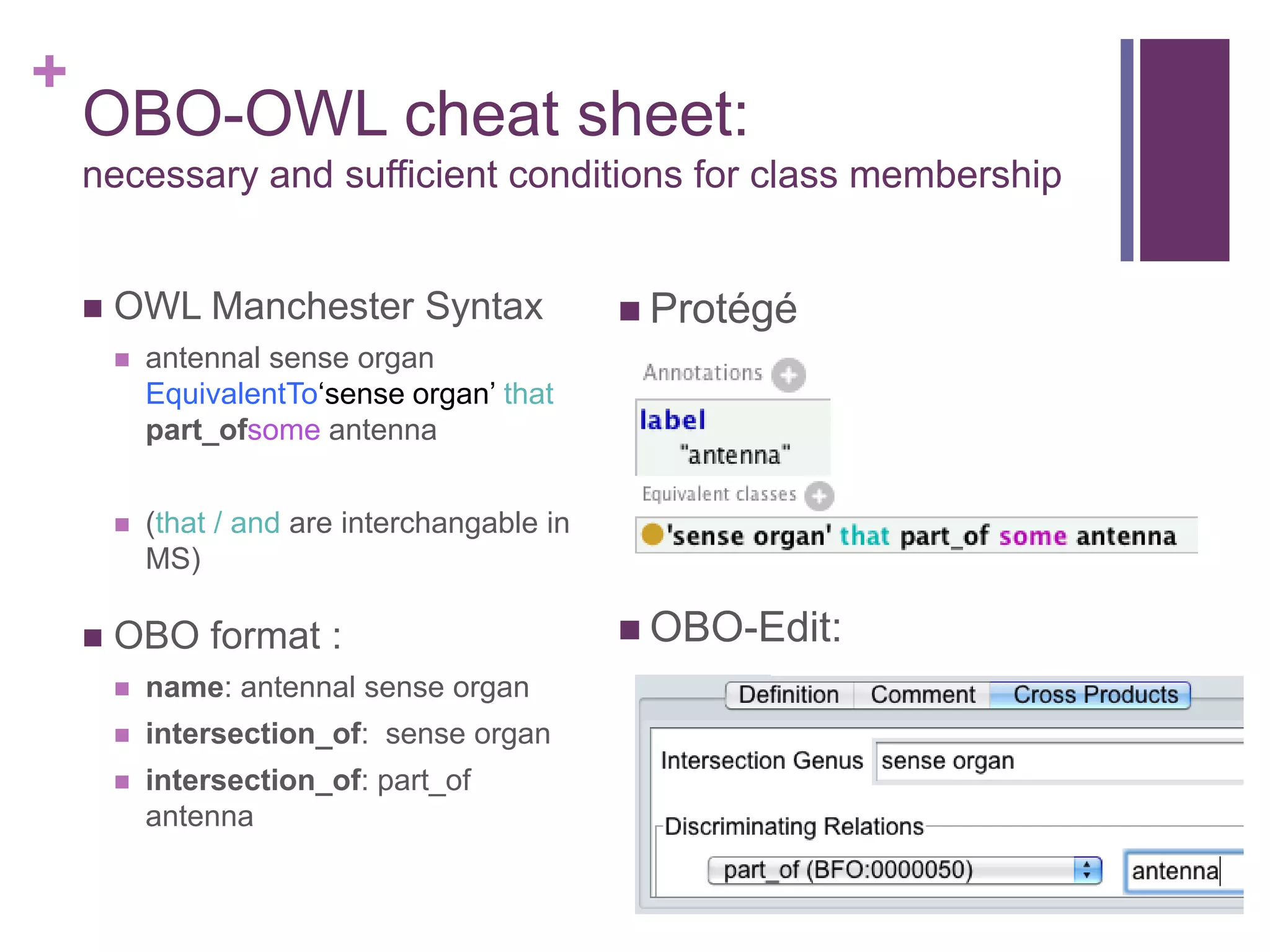
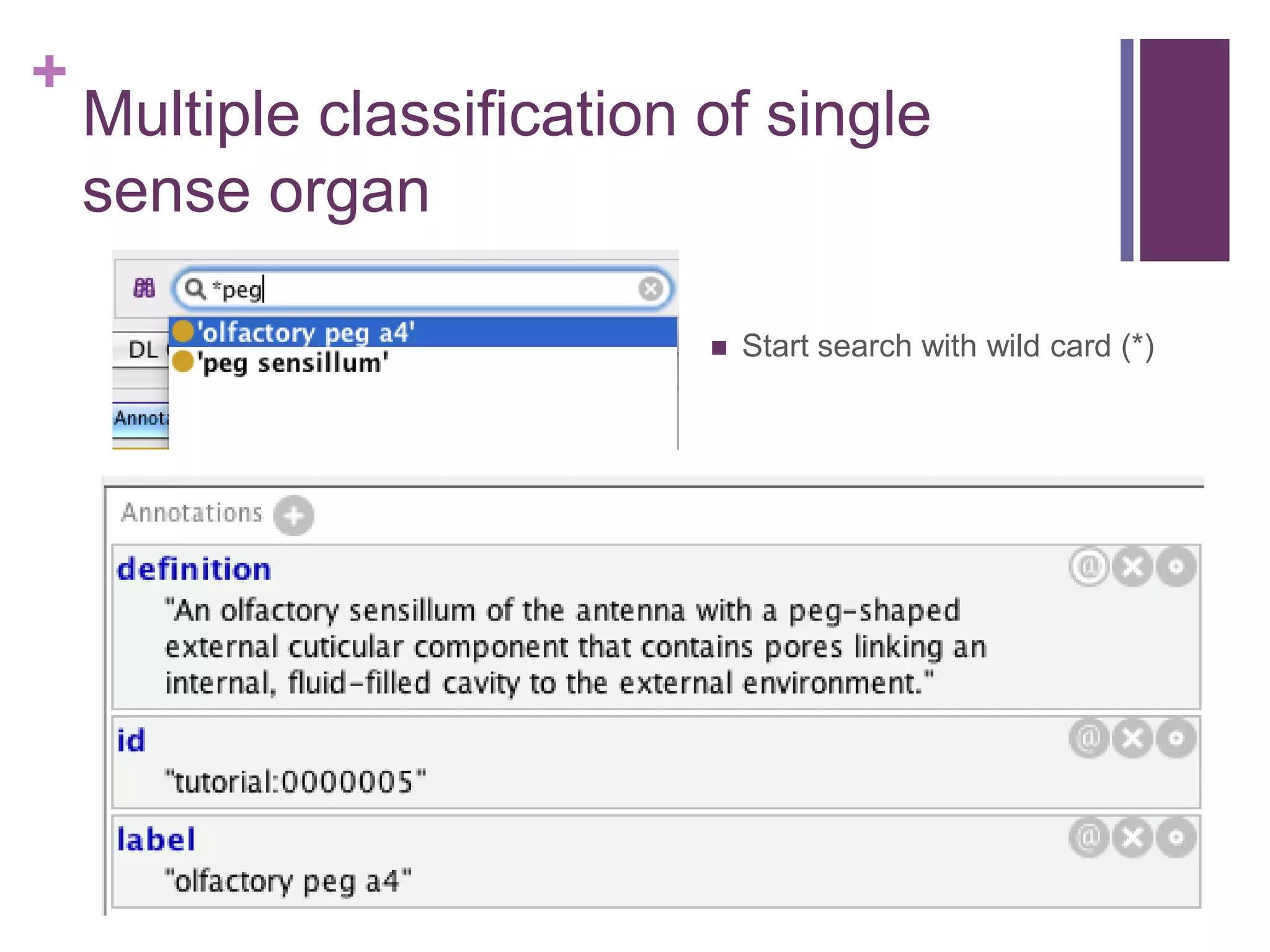
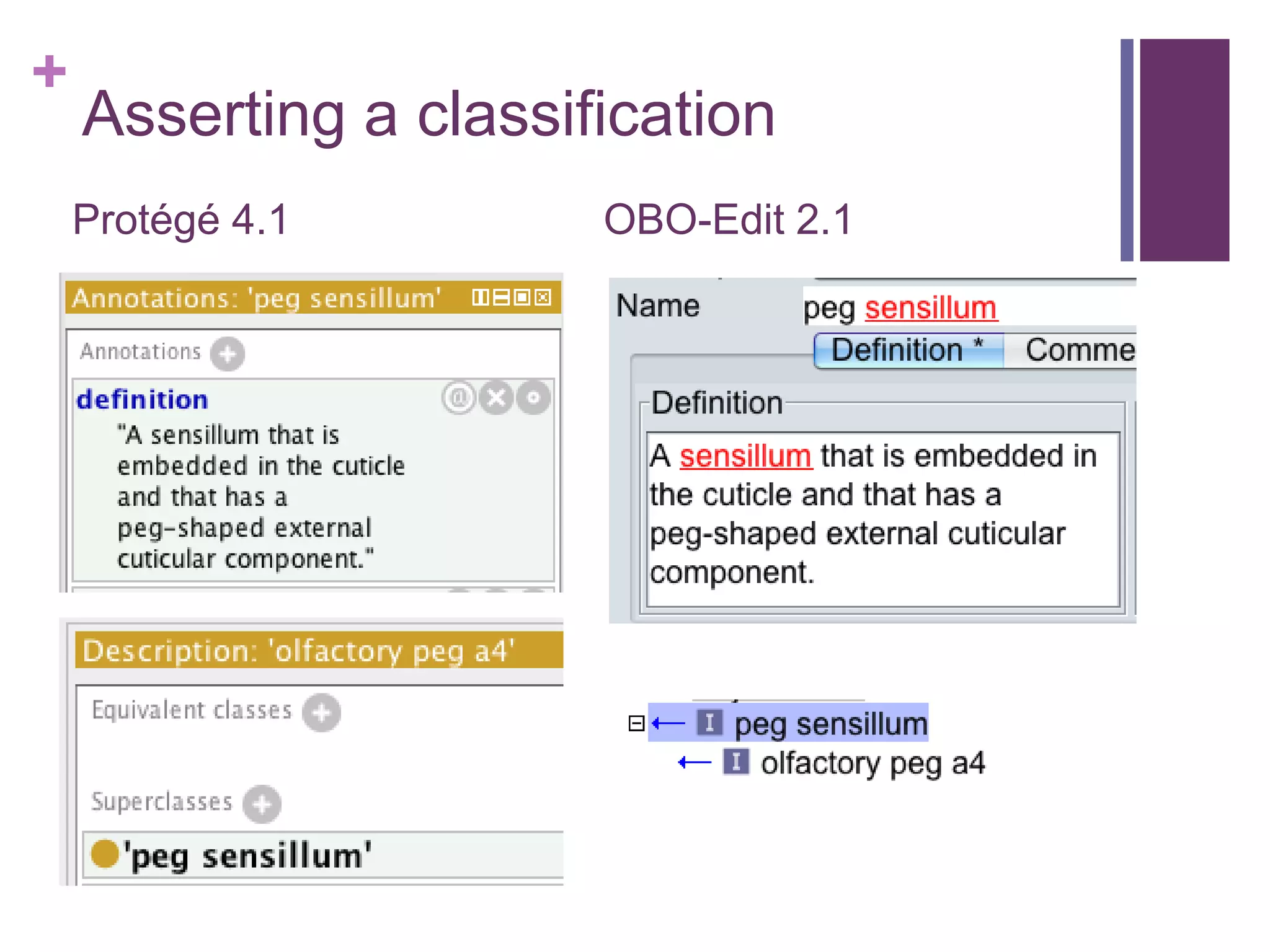
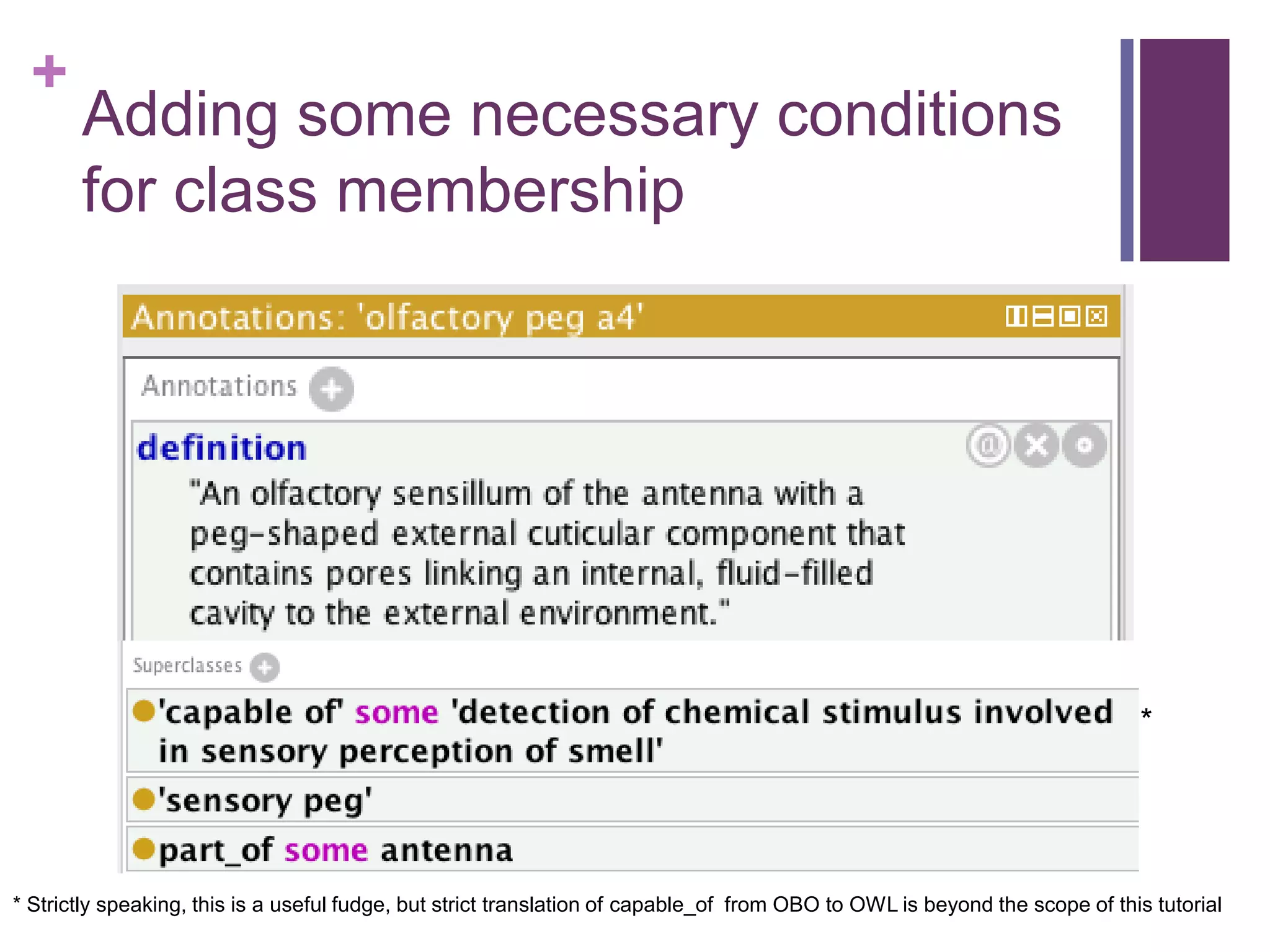
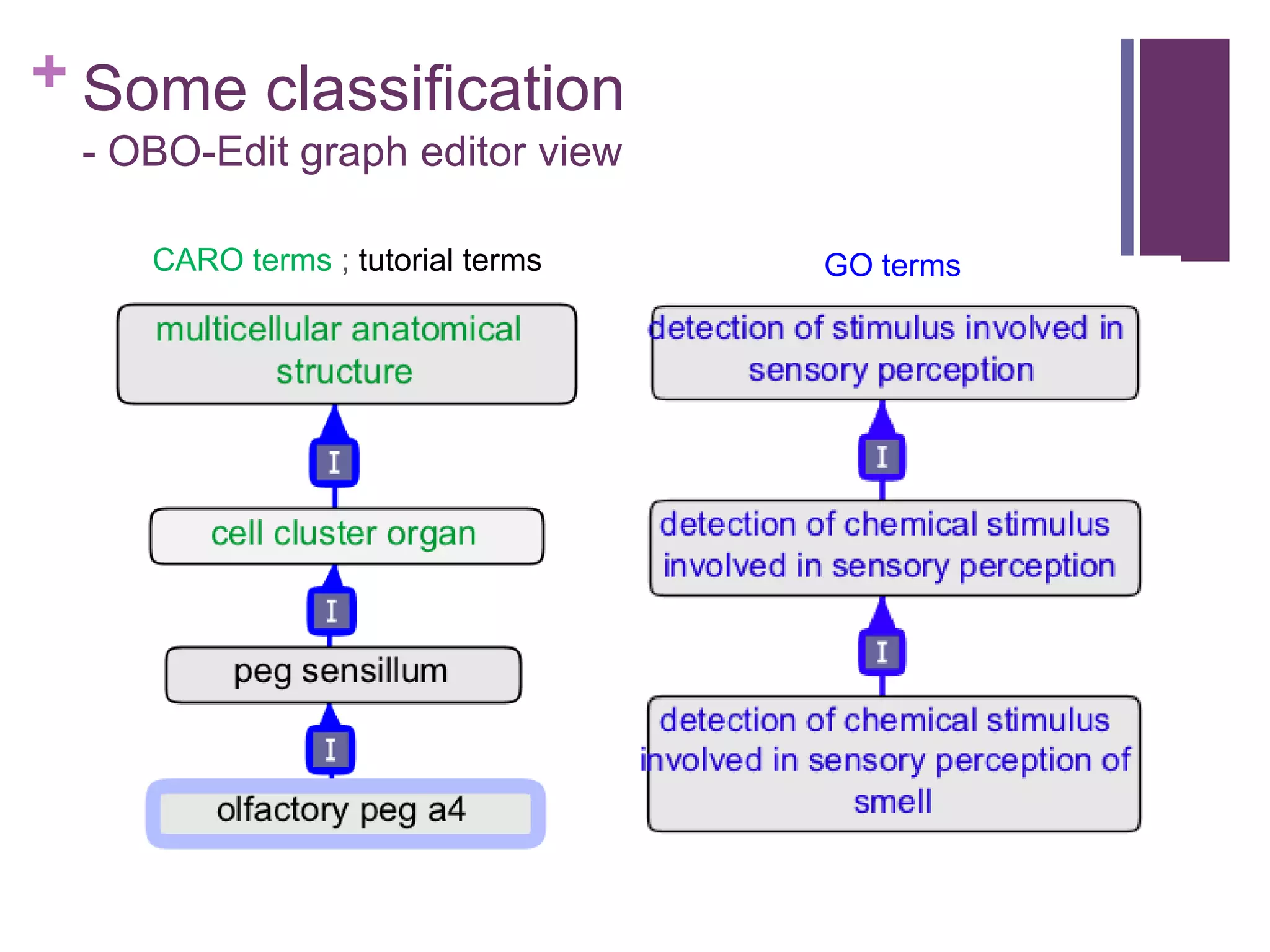
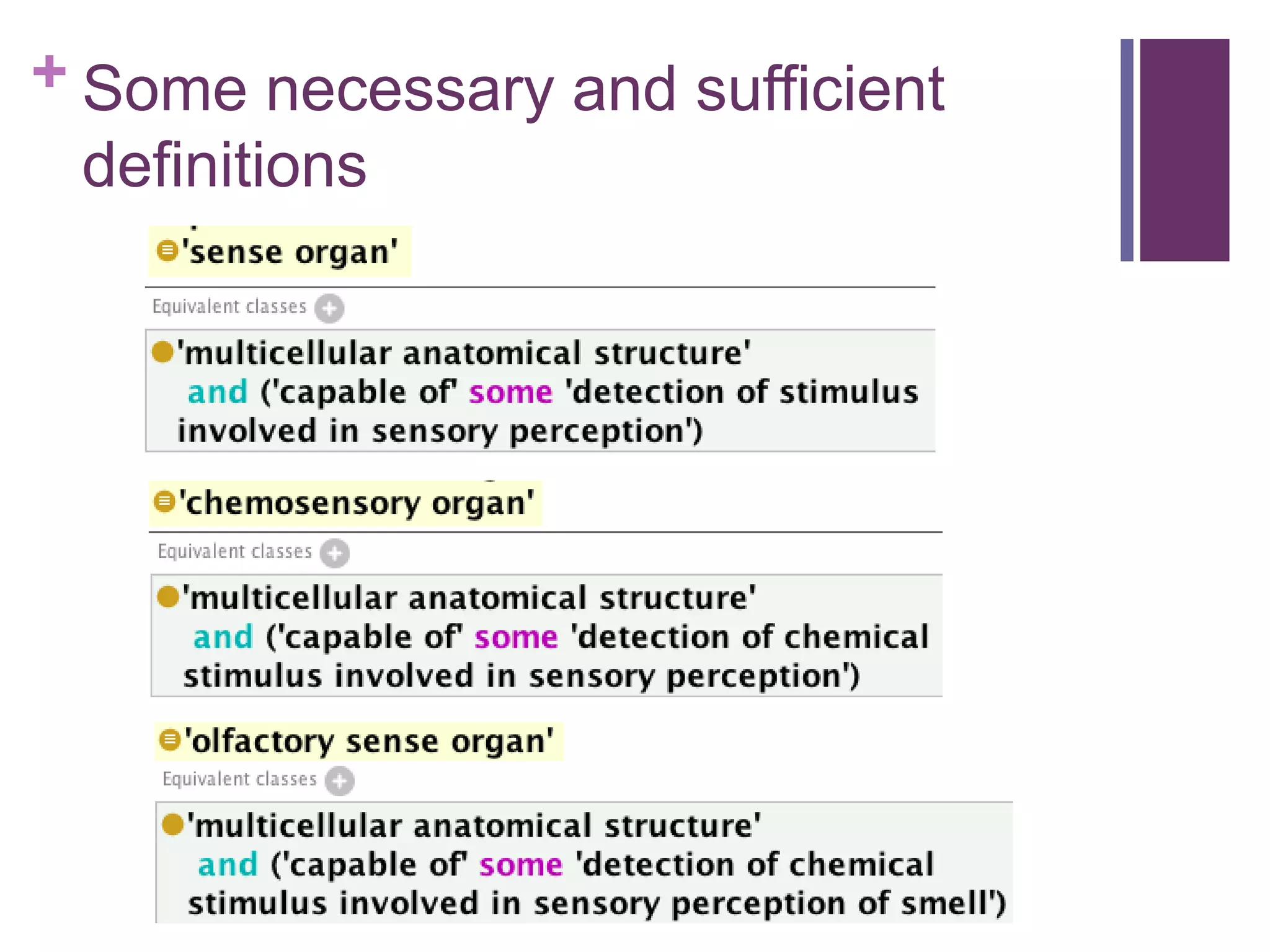
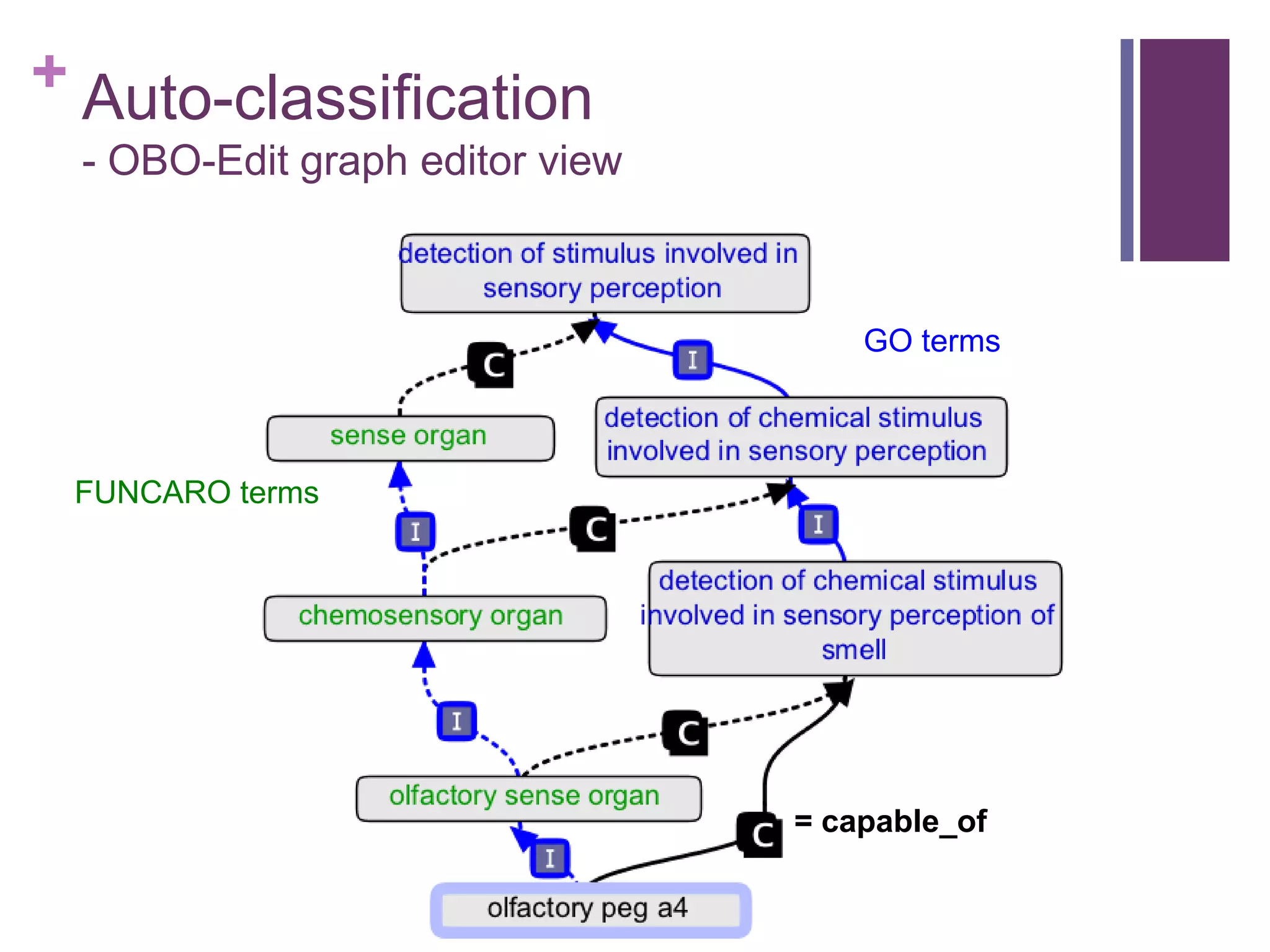
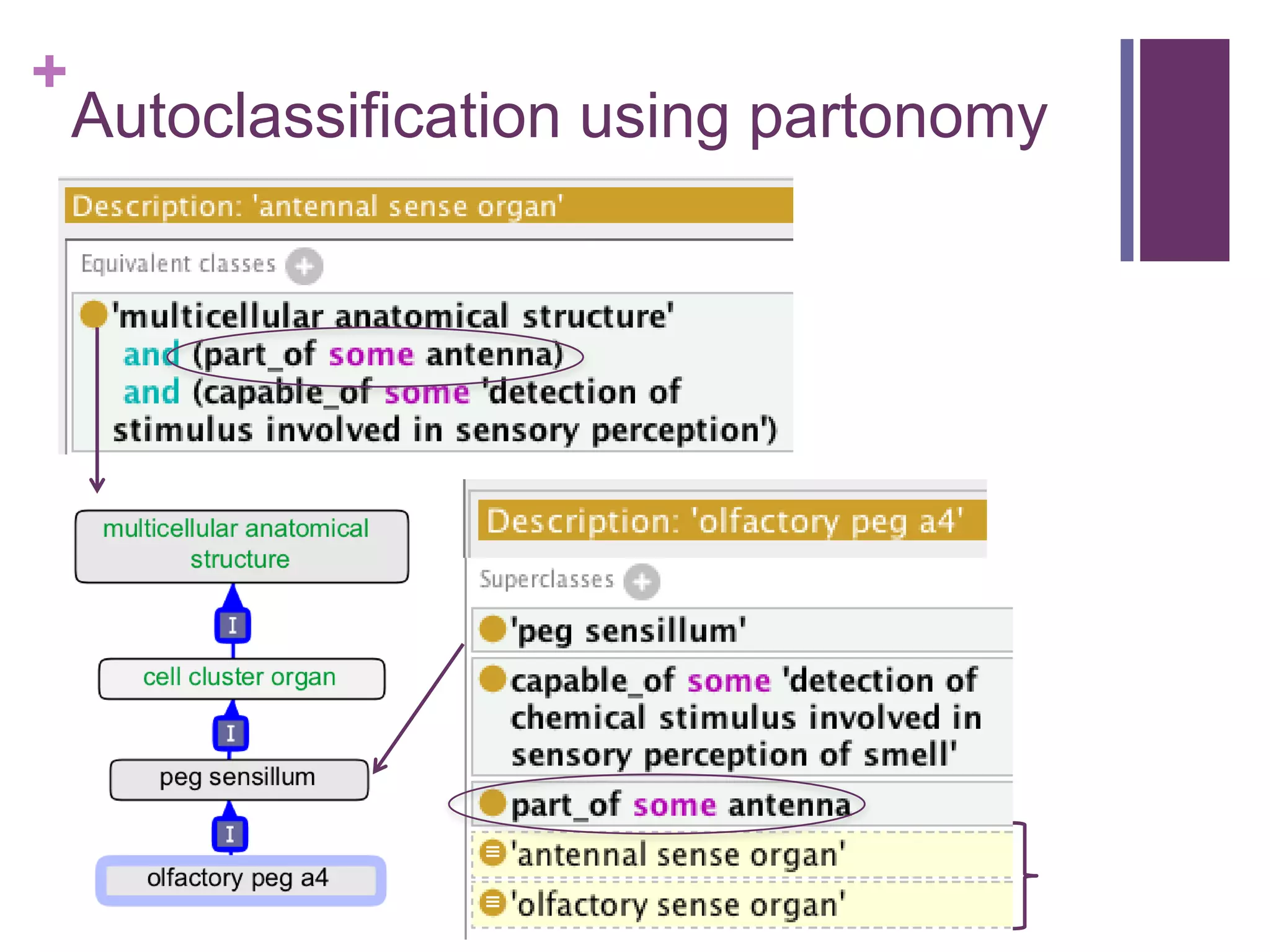
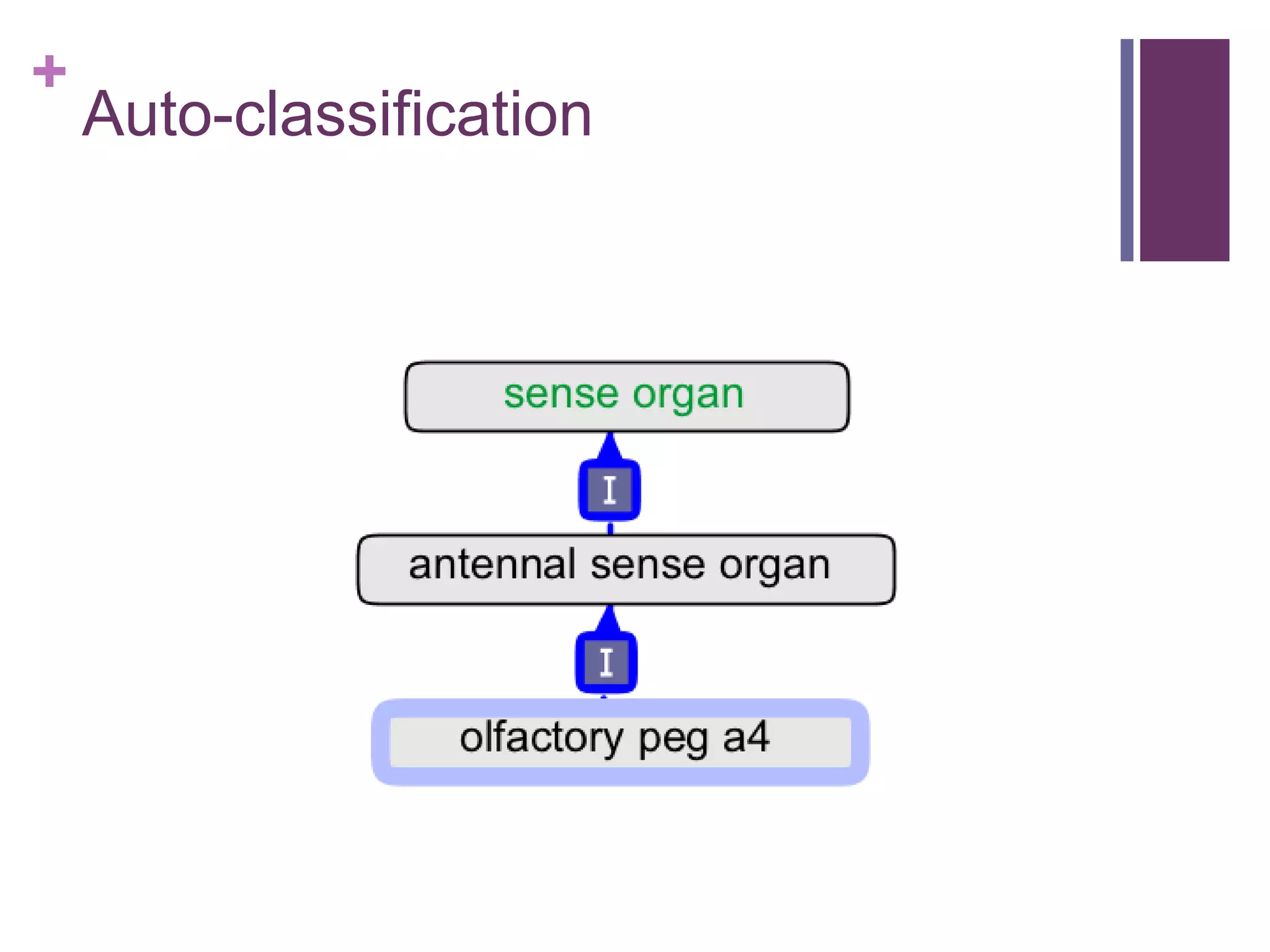
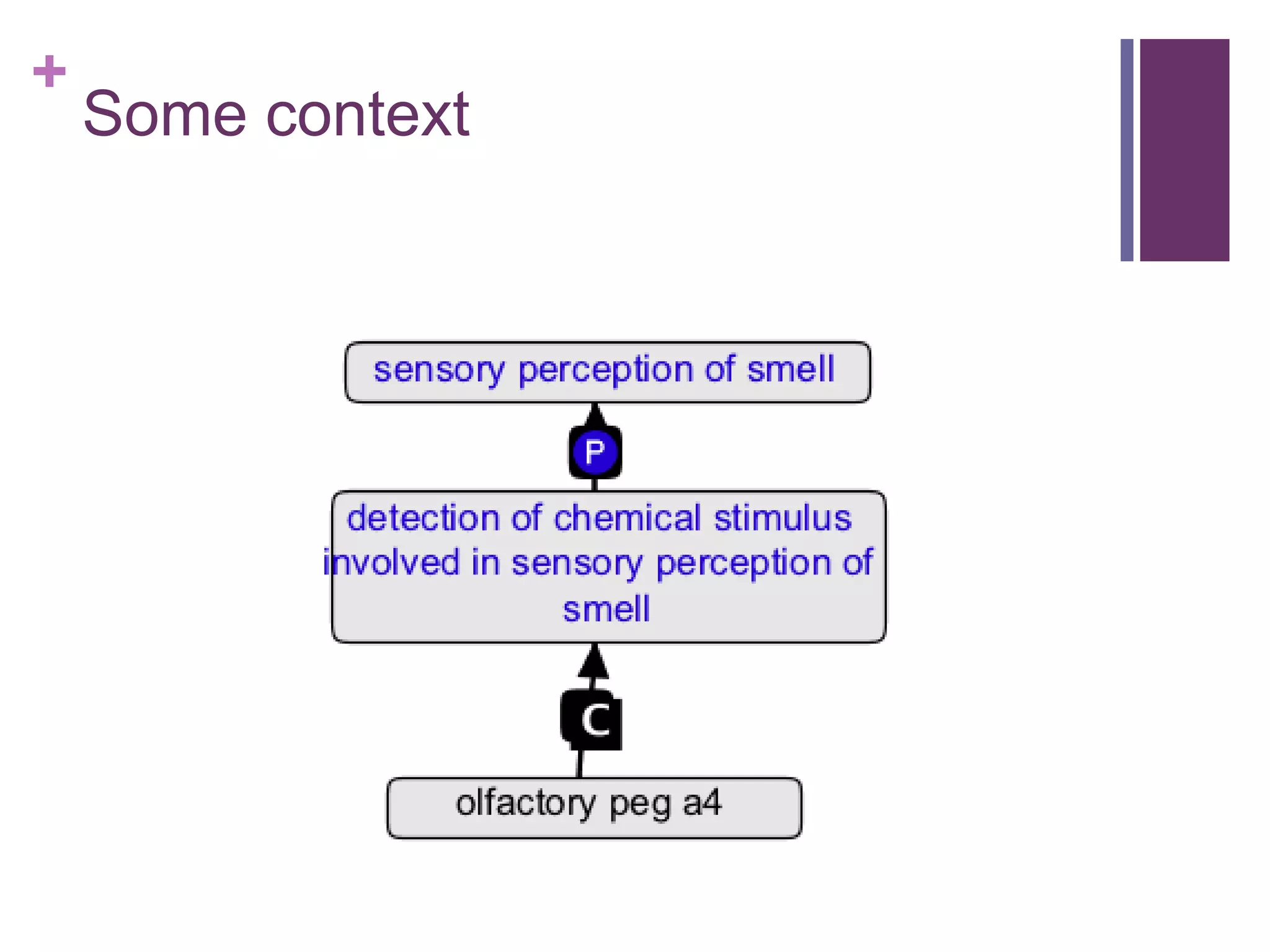
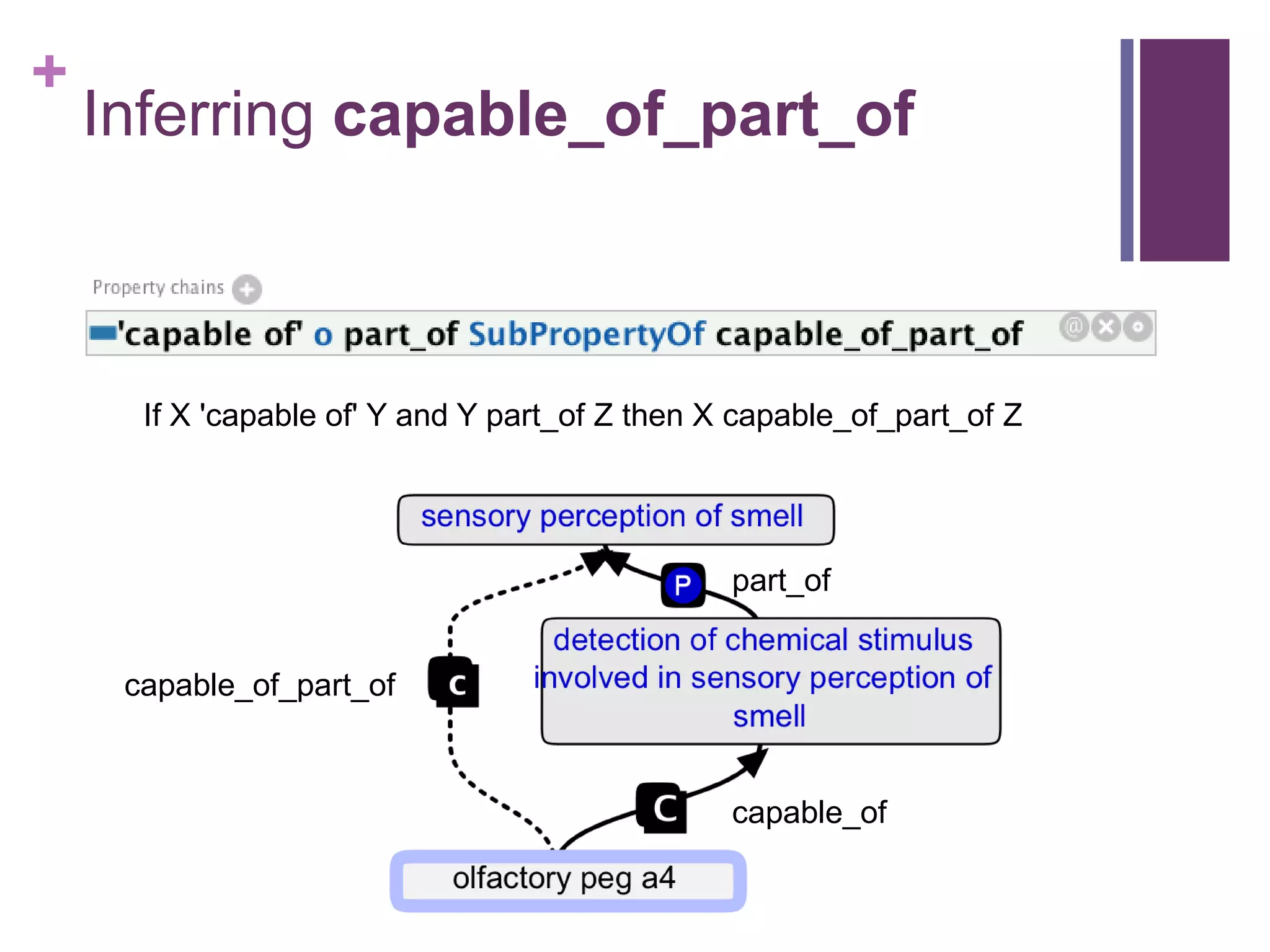
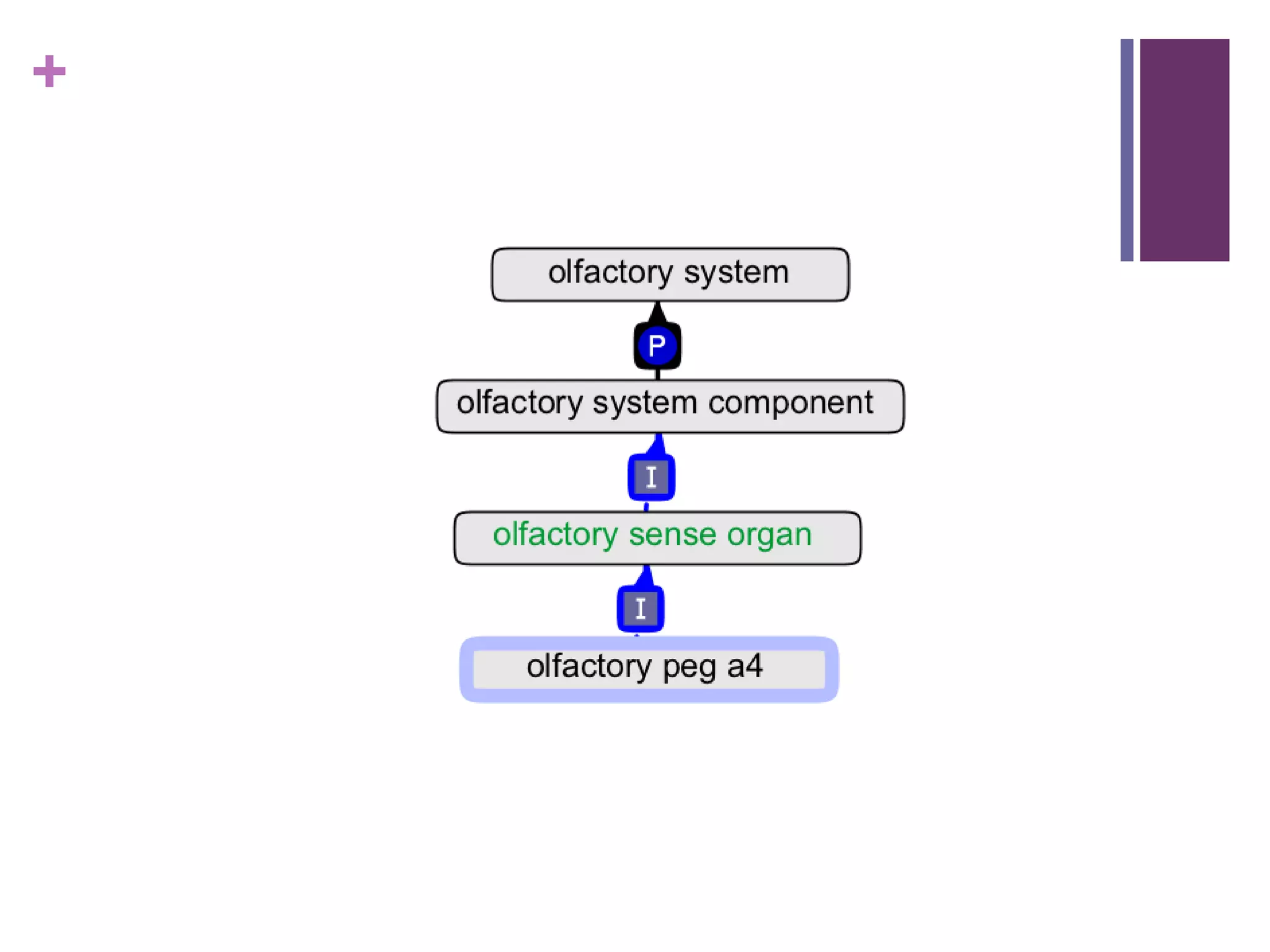
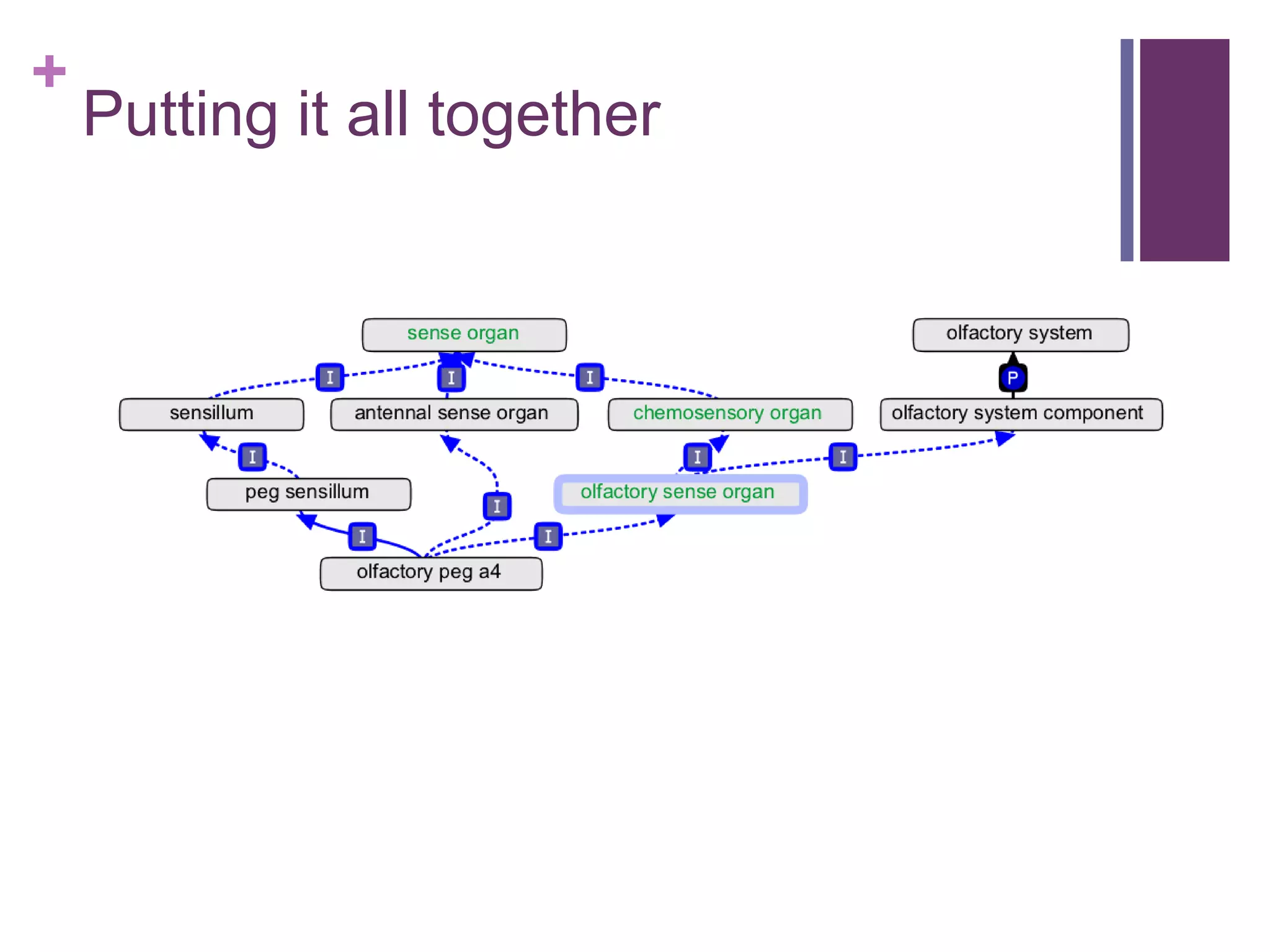
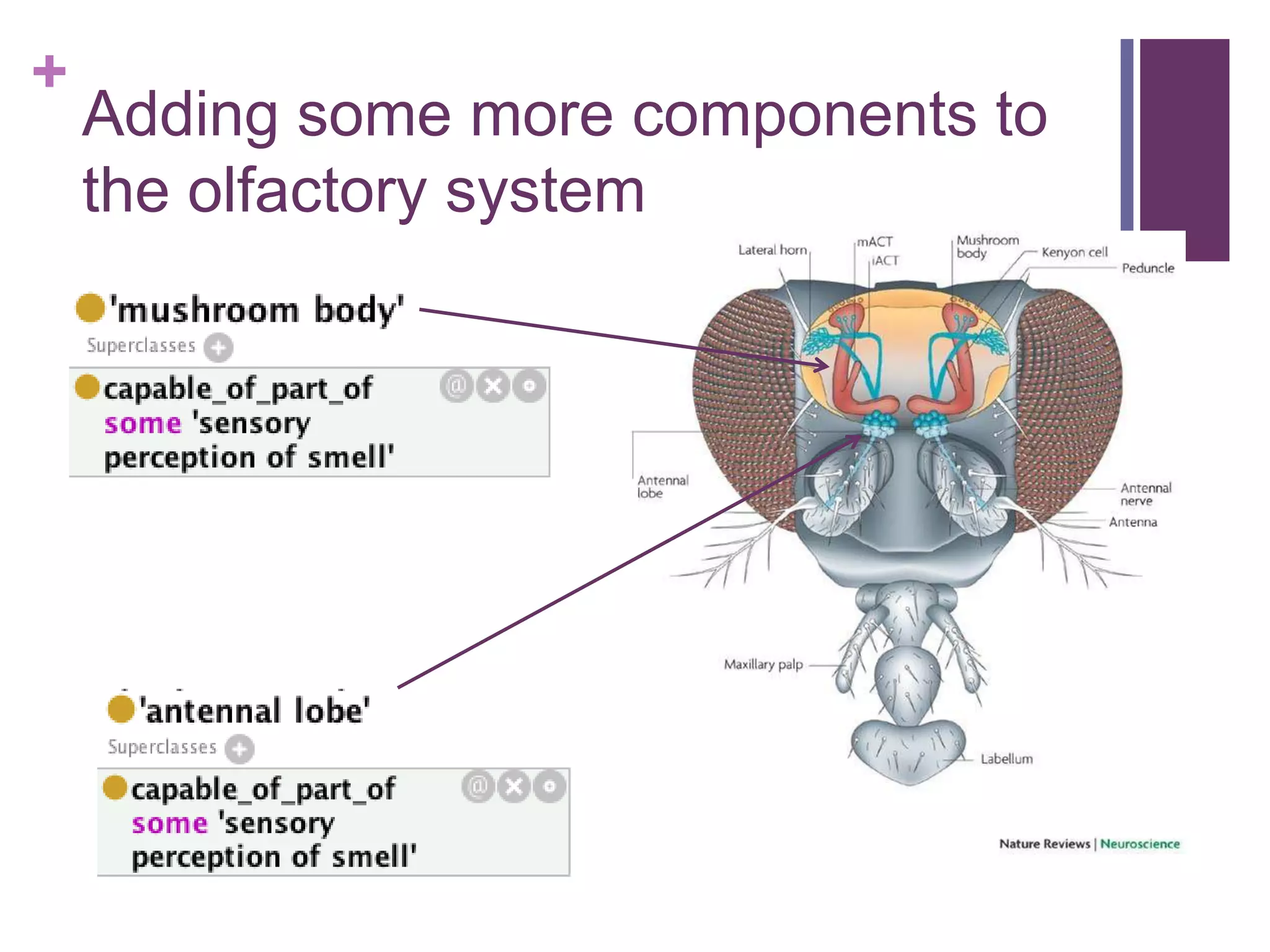
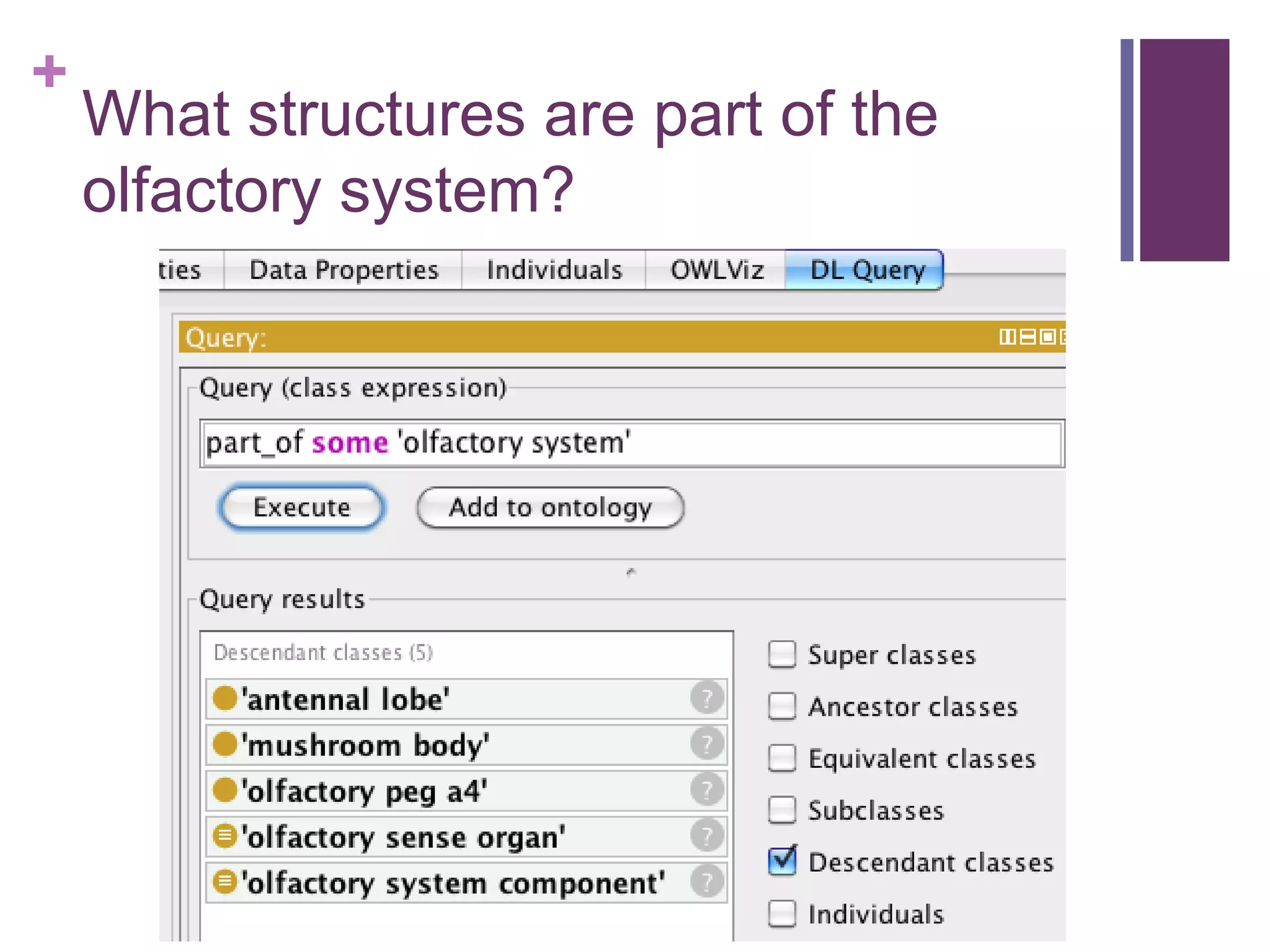
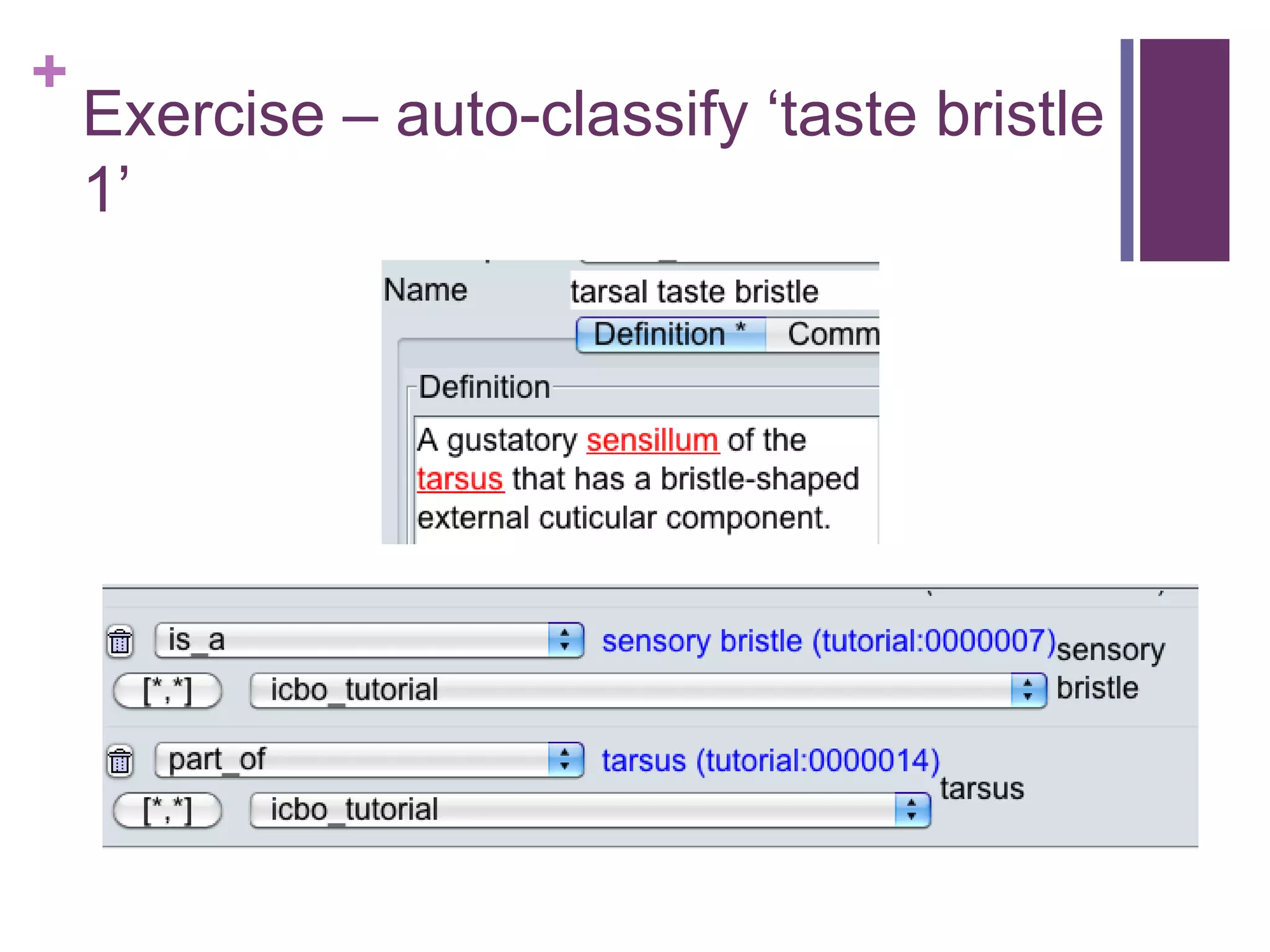
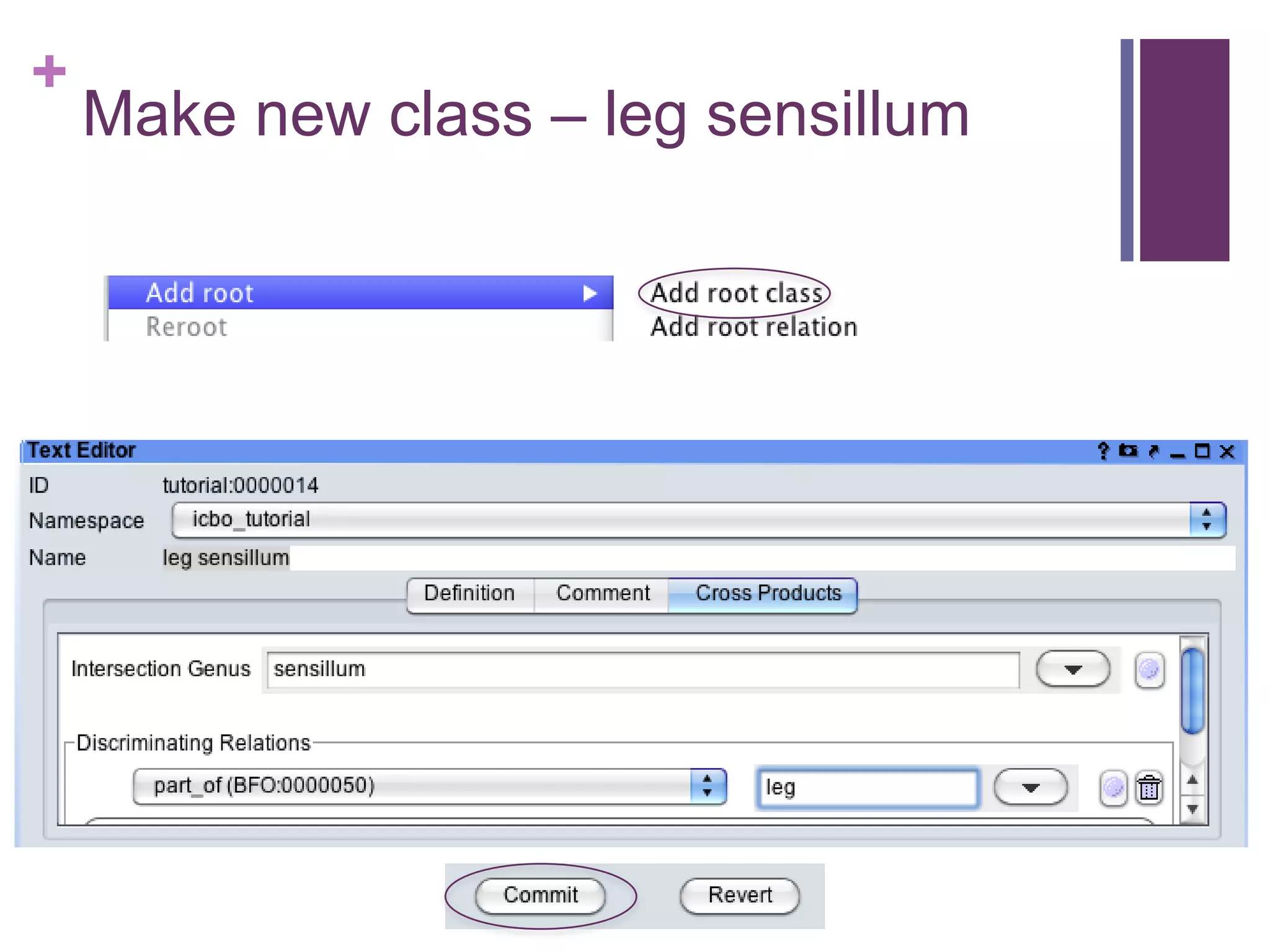
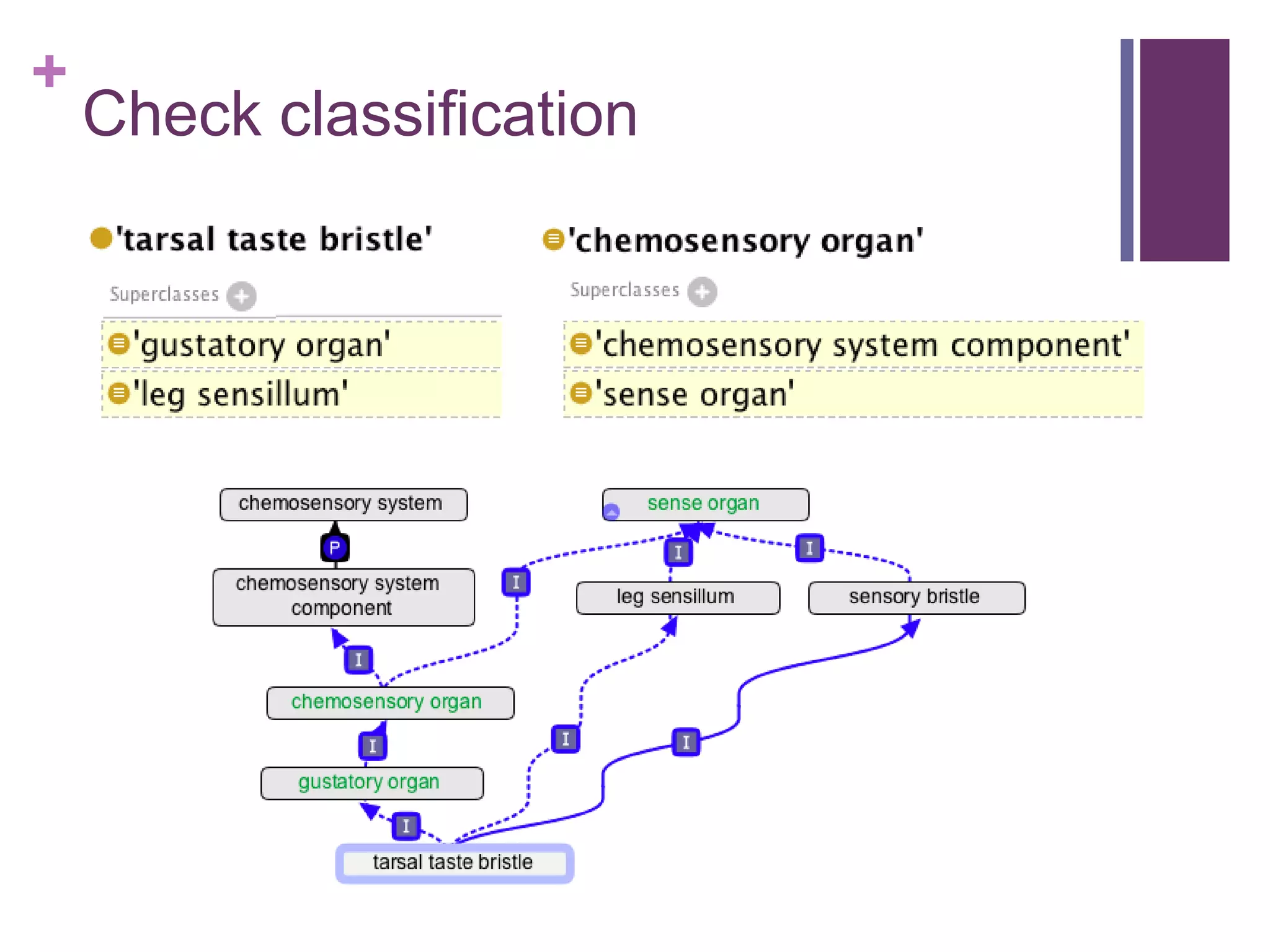
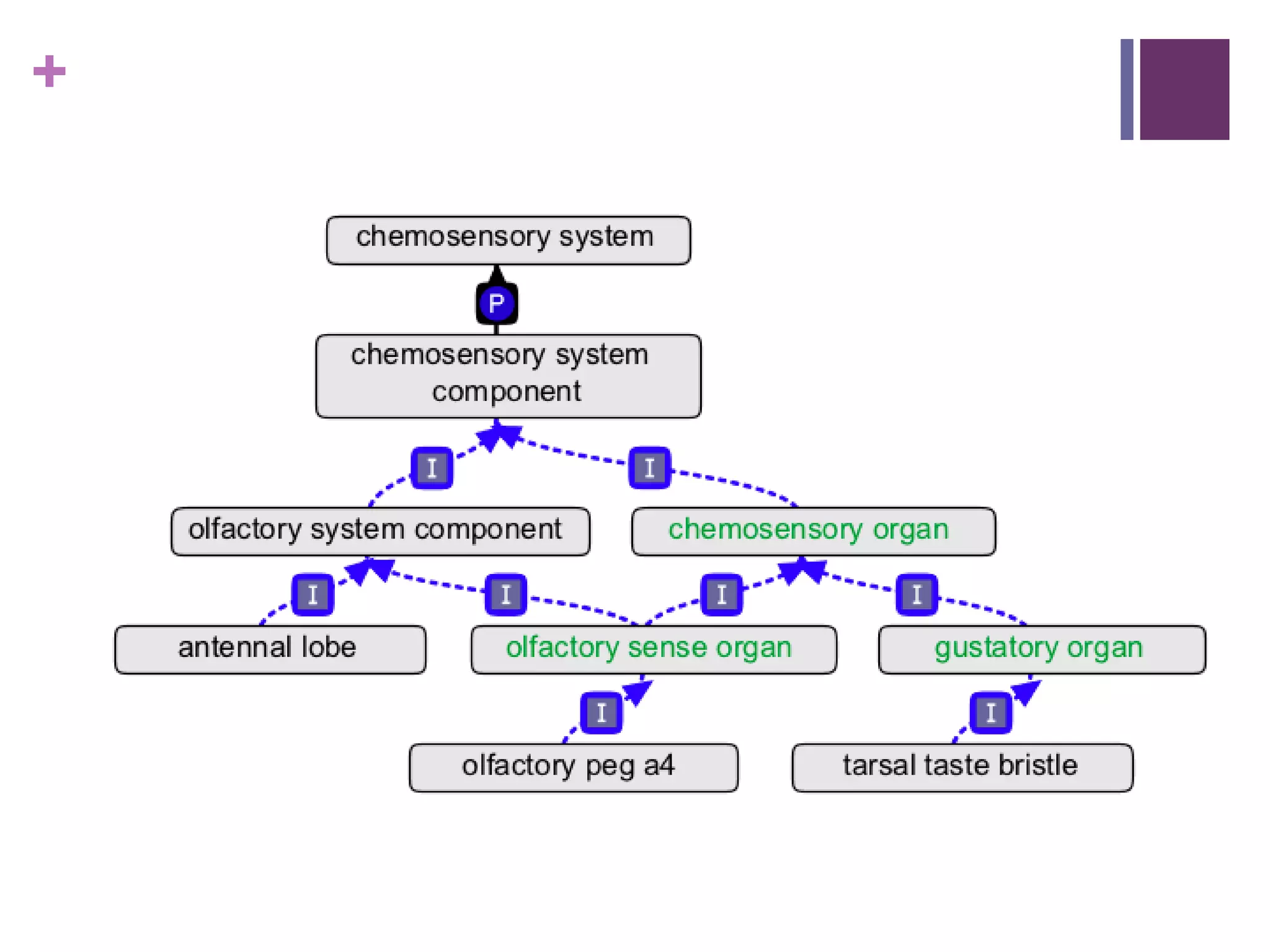
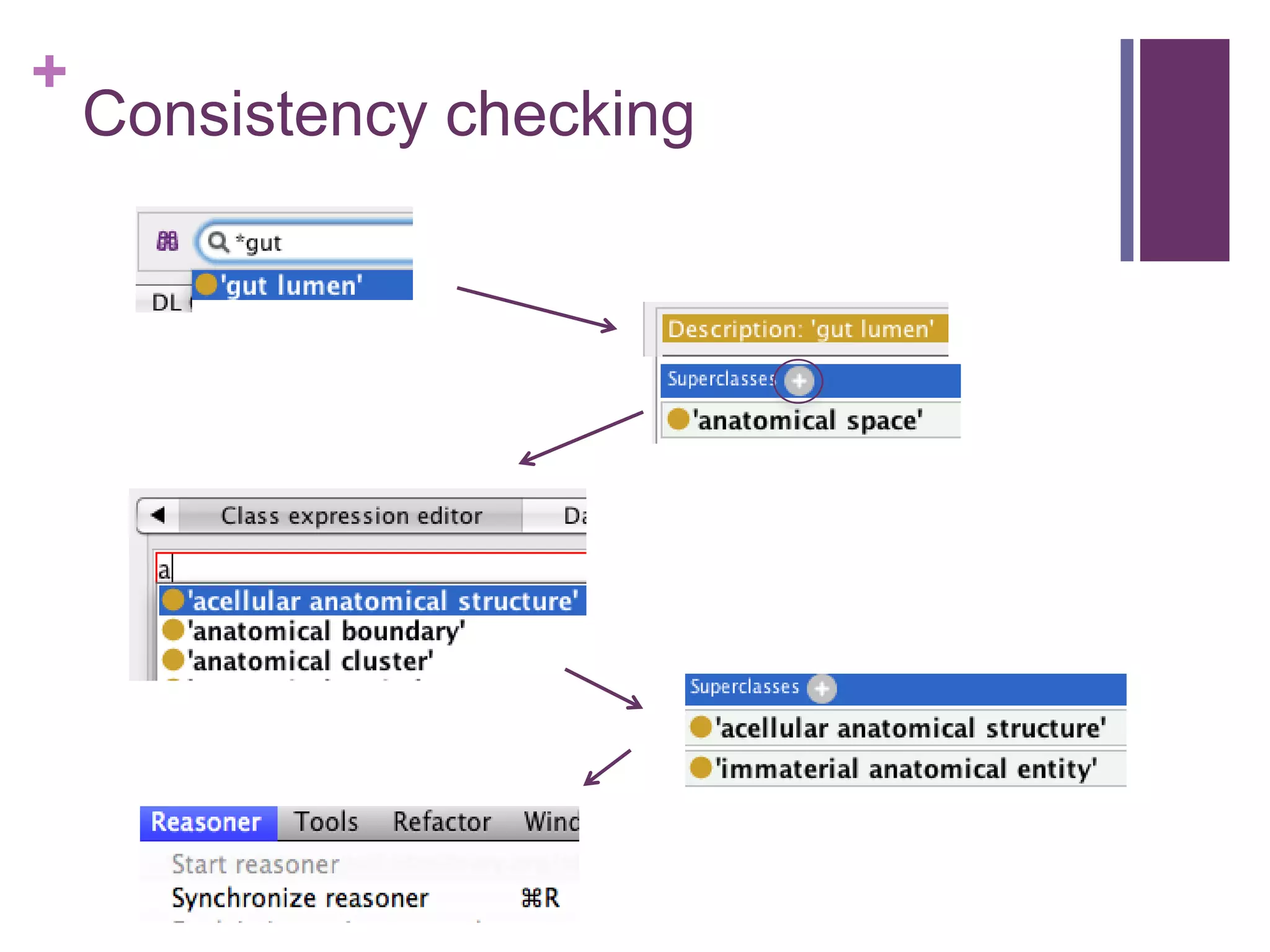
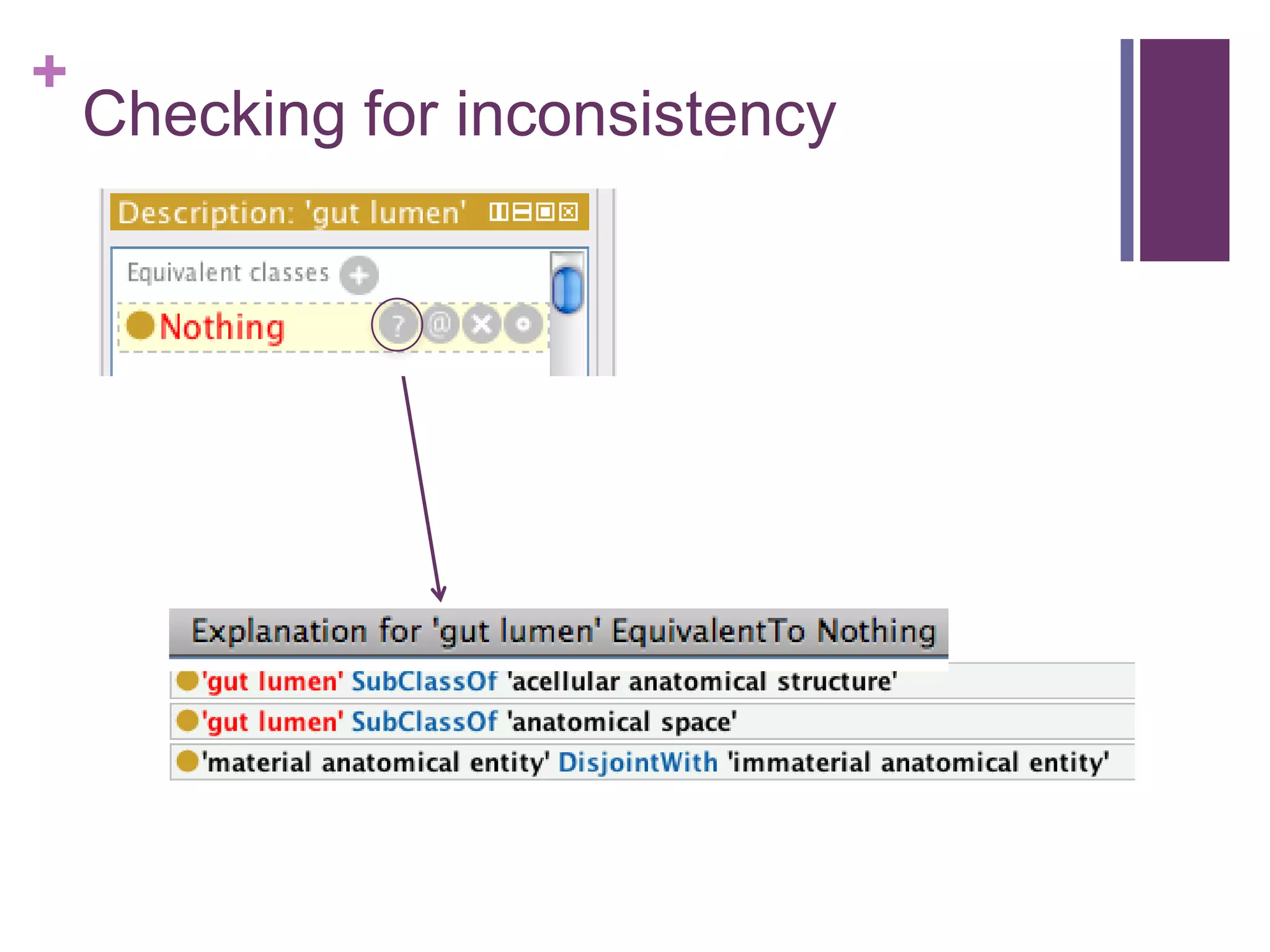
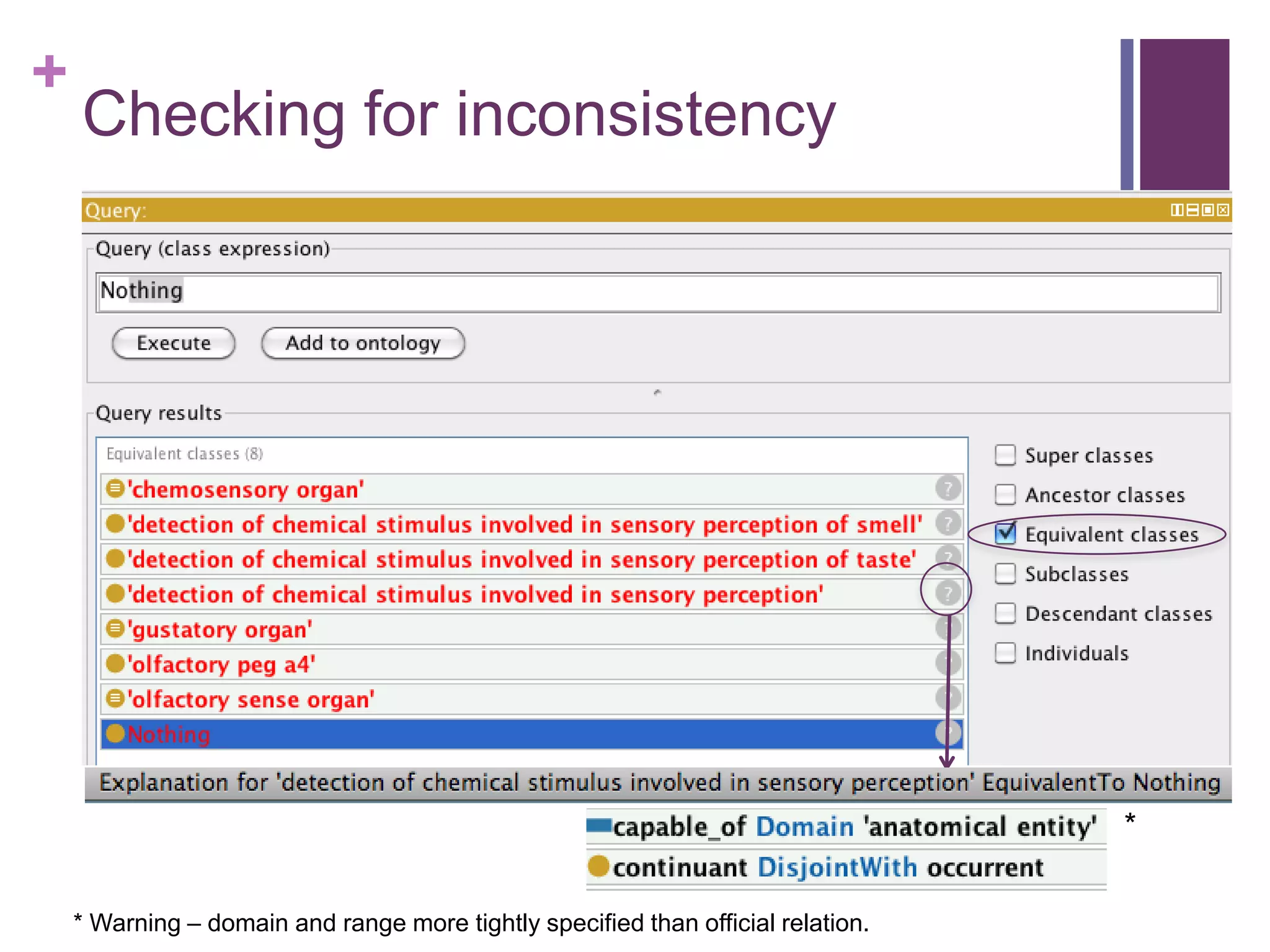
This document provides an introduction to converting ontologies between the OBO format and the OWL format. It discusses the benefits of using OWL, including taking advantage of reasoning and automated classification. It also introduces Oort, a tool for generating OBO files that do not require reasoning from ontologies that do. The document then provides a tutorial on building ontologies, including maintaining multiple classification schemes, using relationships to specify necessary and sufficient conditions for class membership, and using error messages to identify issues.









![+
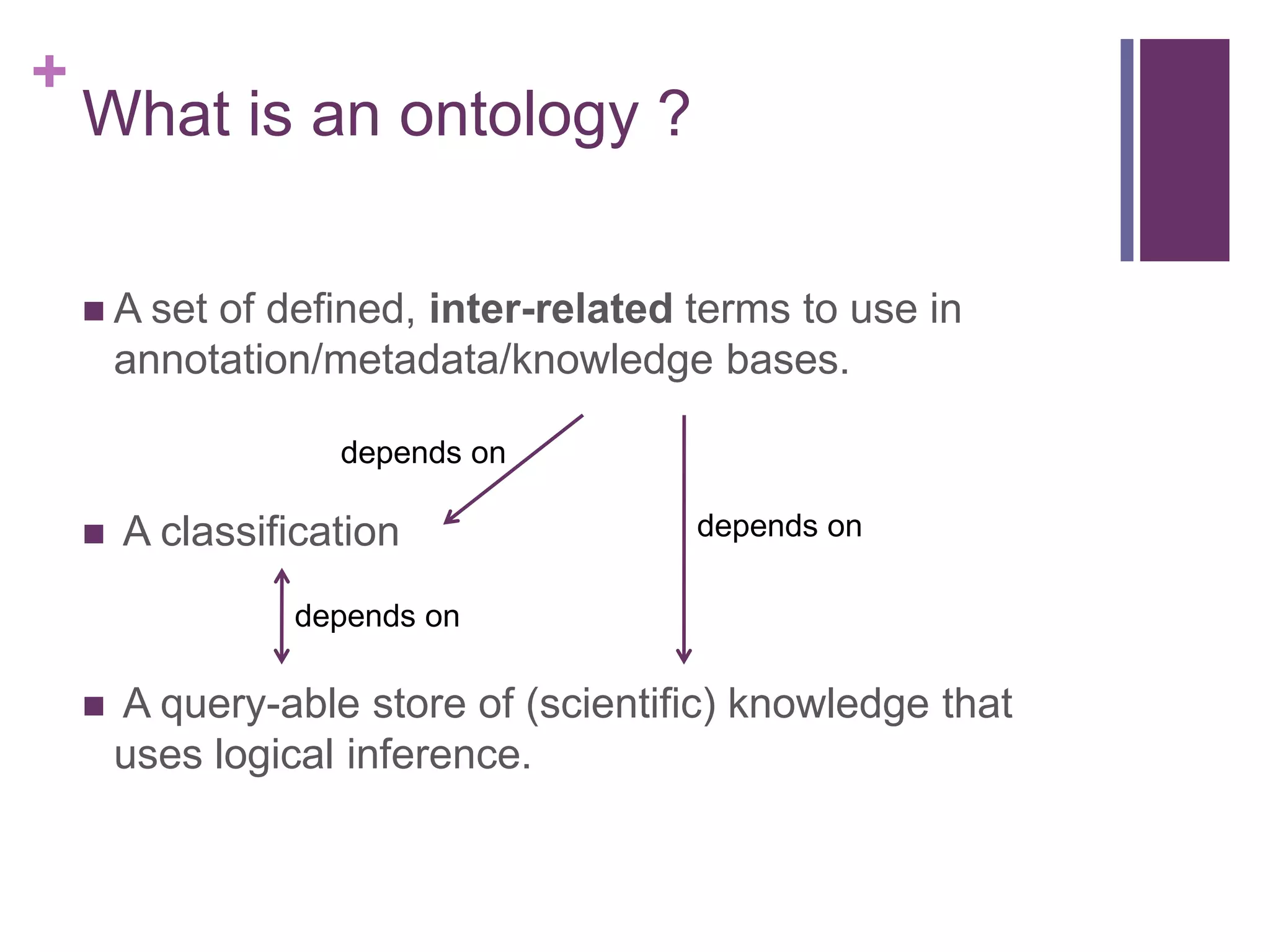
What is an (OBO-foundry) ontology ?
An ontology contains terms
Terms refer to (denote) types
(classes)
Types are classifications of things
(instances) in the real world, based on
some set of criteria.
My left hand is an instance of the type hand
The criteria for class membership is
recorded using textual definitions, at
least some elements of which are
formalized as relationships.
name: hand
def: “An anatomical structure that has four
fingers and a thumb and is attached to the
end of an arm.” [reference: DOS]
relationship: hand has_part finger
relationship: hand has_part thumb
relationship: part_of arm
Image from Gray’s Anatomy (copyright expired)](https://image.slidesharecdn.com/tutorialslides3-110730094944-phpapp02/75/From-OBO-to-OWL-and-back-building-scalable-ontologies-12-2048.jpg)