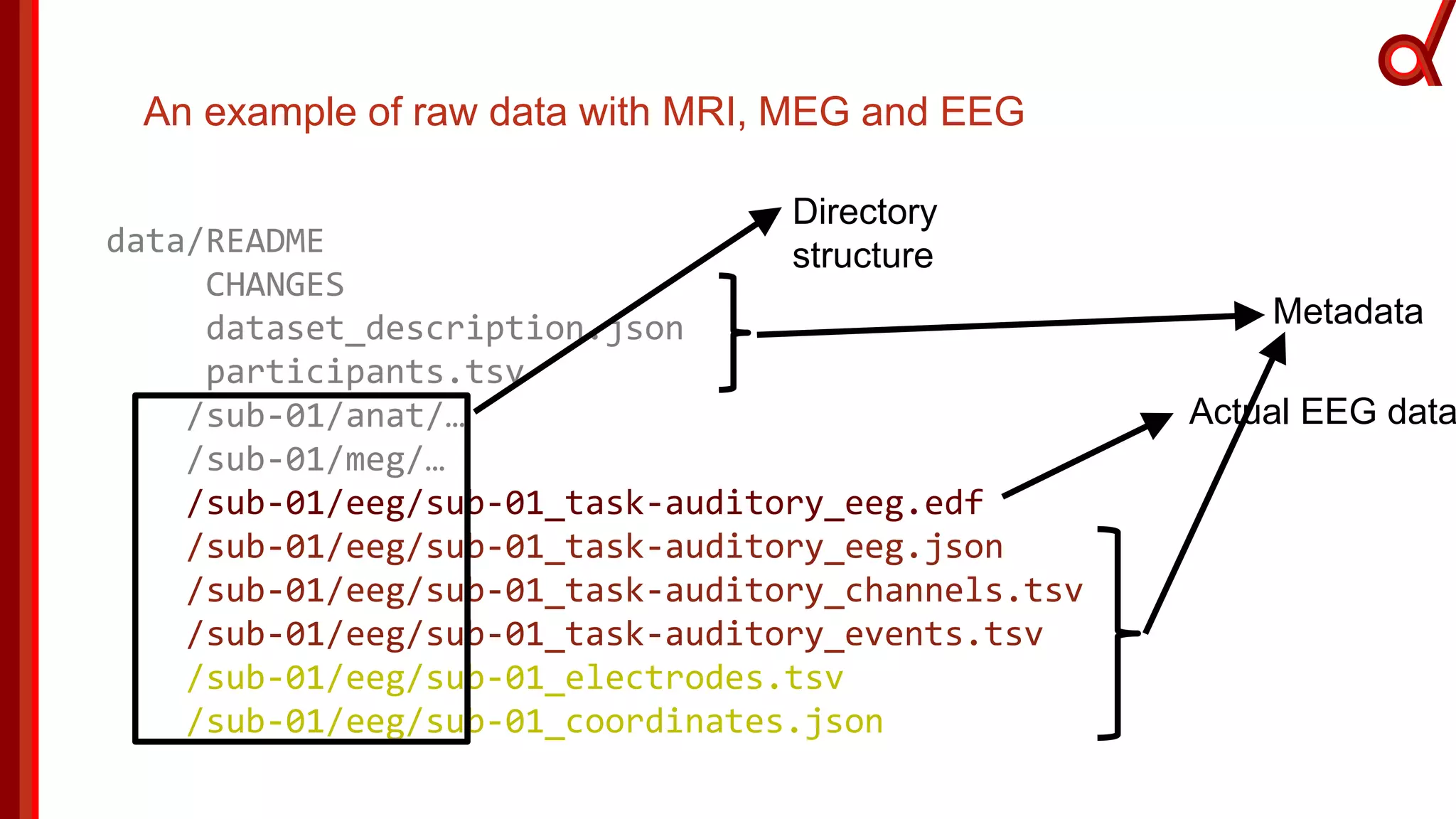
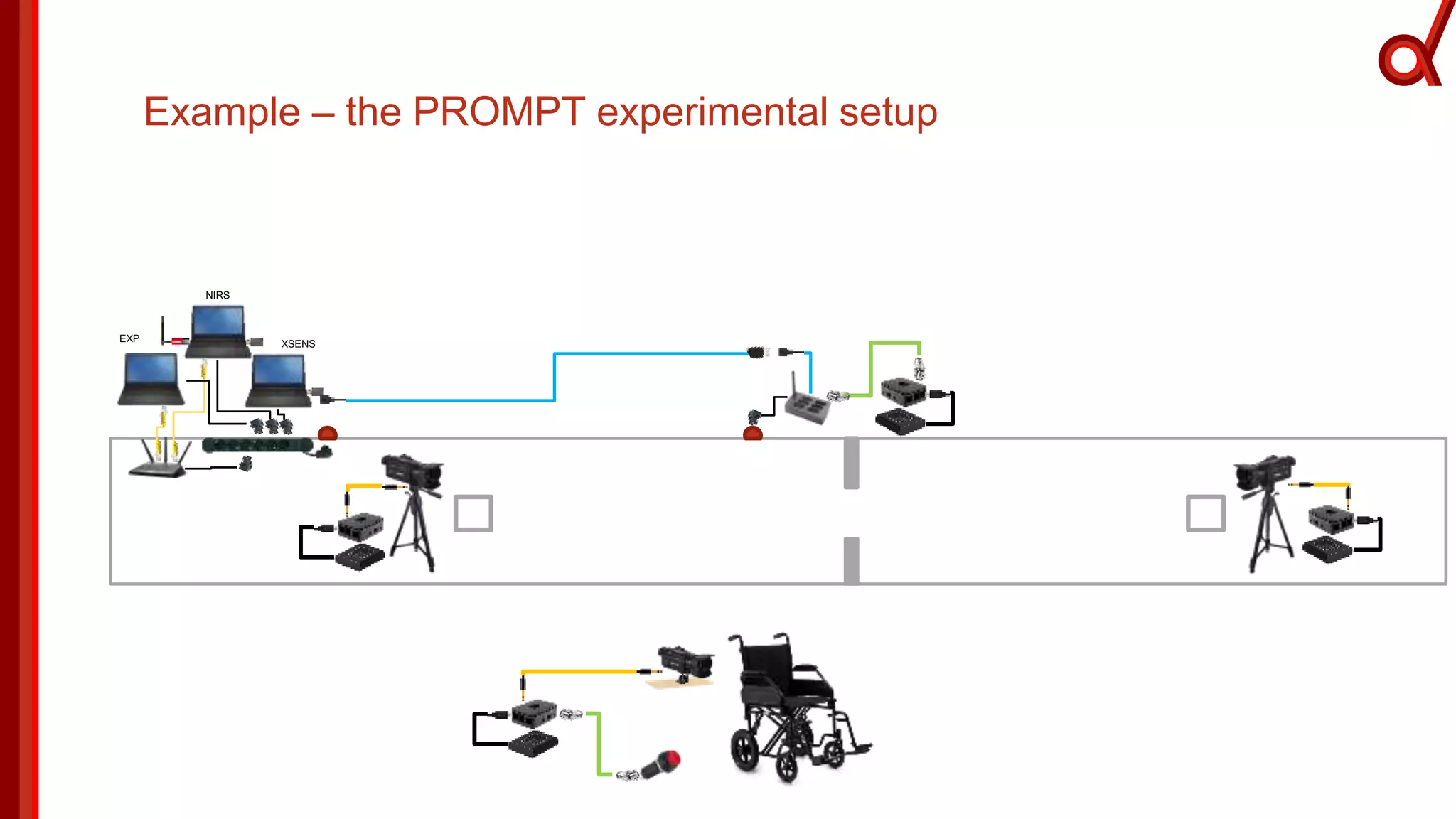
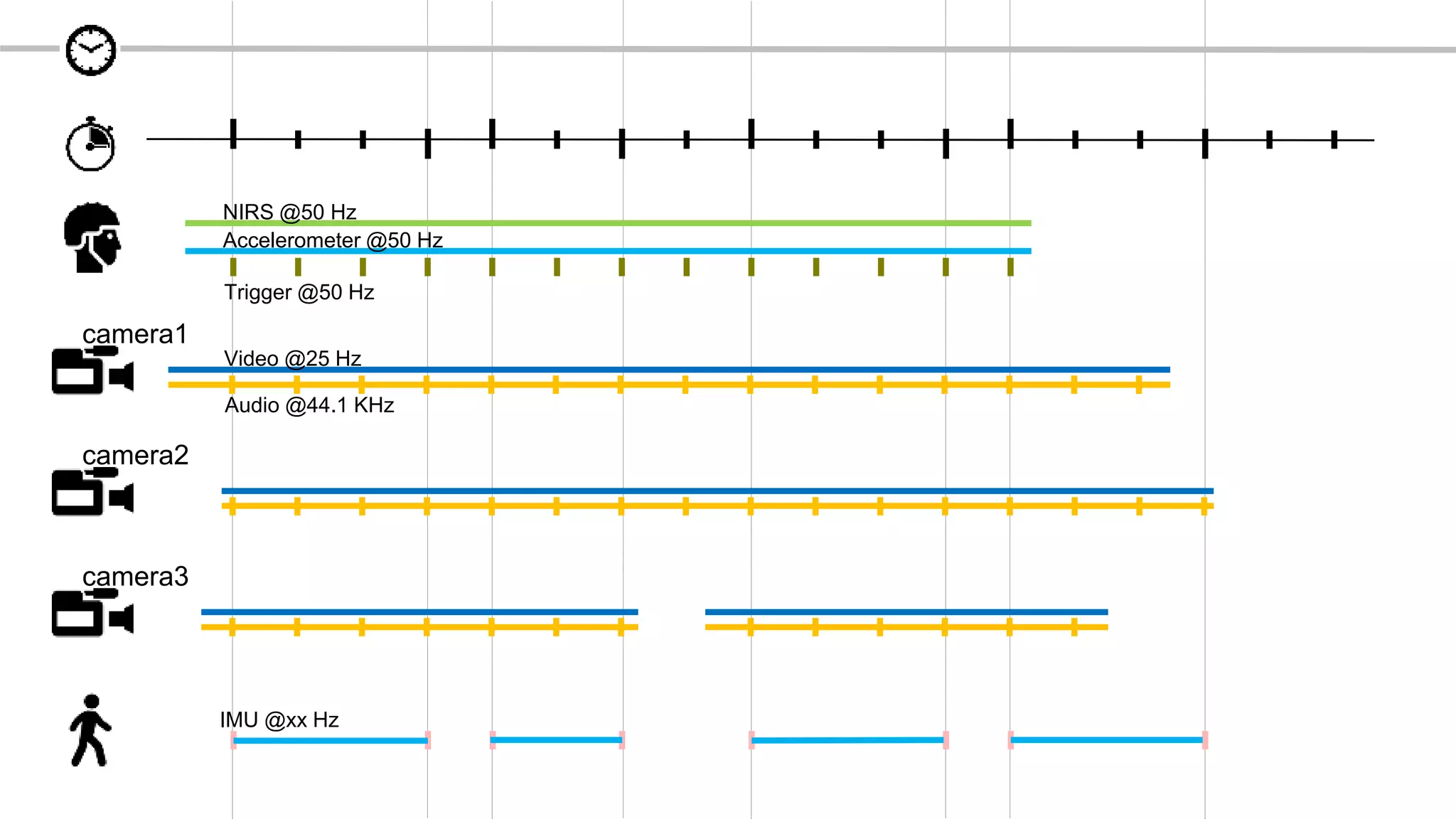
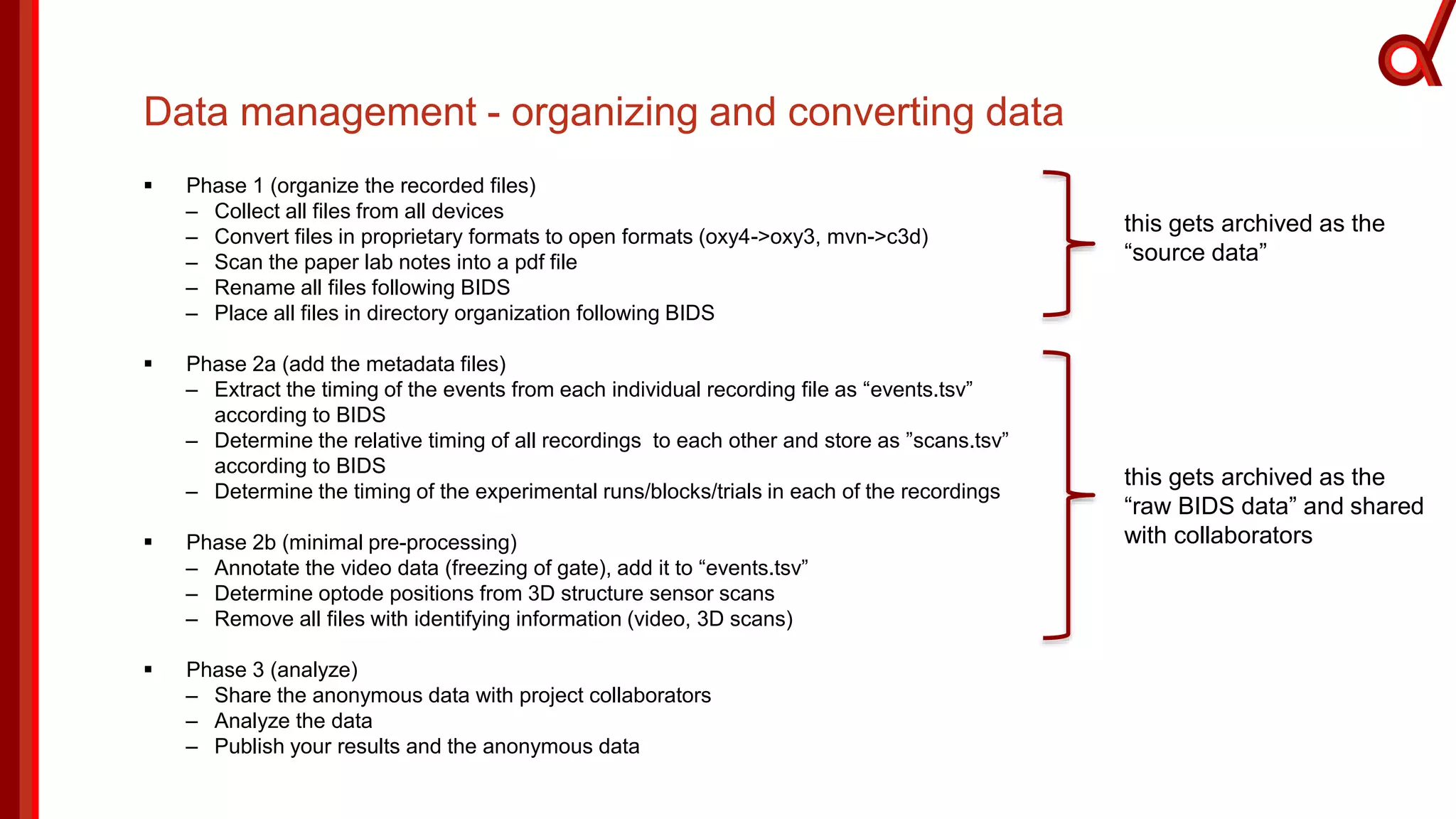
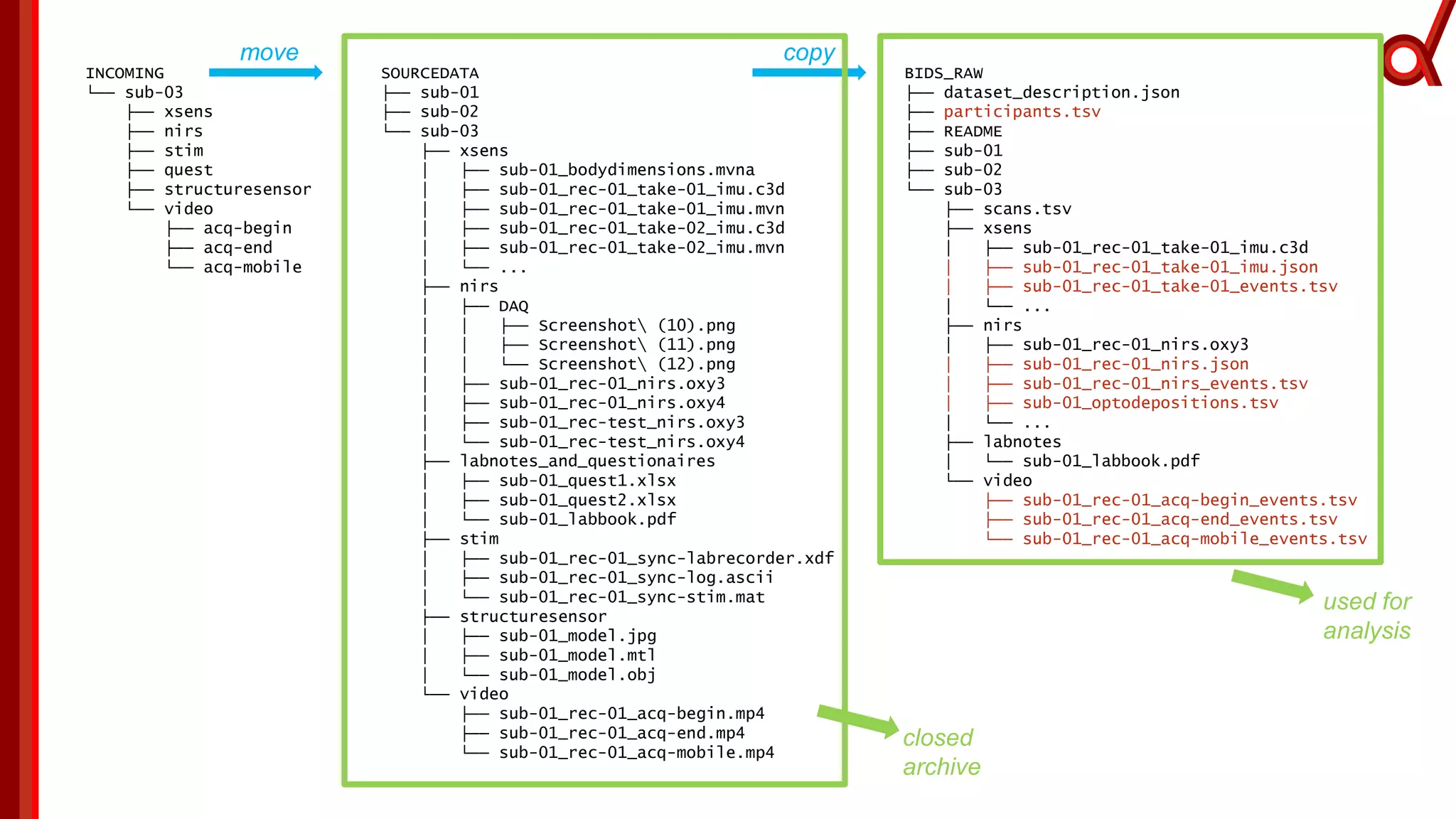
The document discusses the Brain Imaging Data Structure (BIDS), a standardized method for structuring neuroimaging data which addresses metadata retention and improves data interoperability. BIDS has expanded to include various neuroimaging modalities, including fNIRS, and is designed to facilitate analysis, sharing, and reproducibility in research. The document emphasizes the importance of following BIDS guidelines for effective data organization and management in fNIRS research.