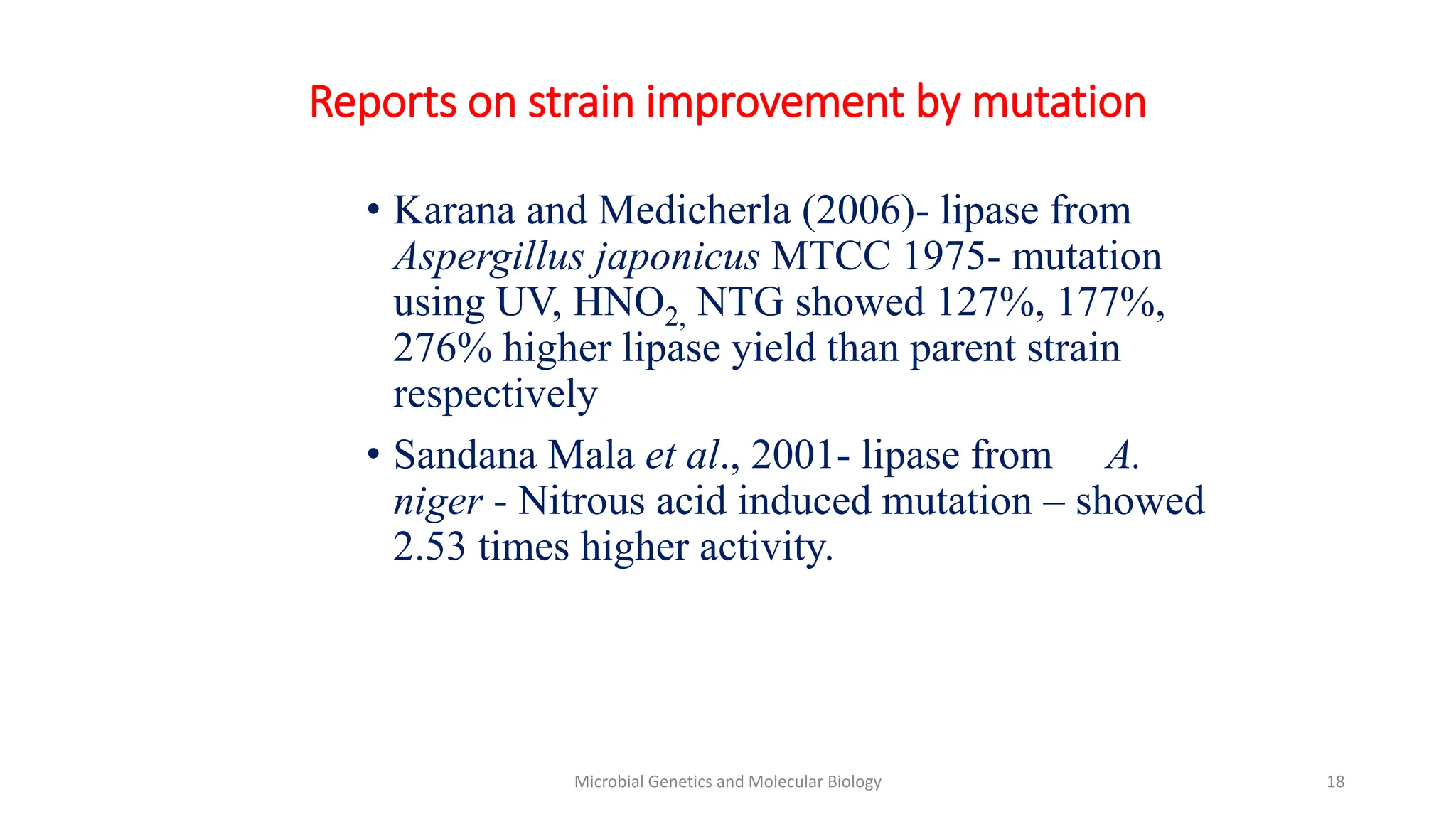
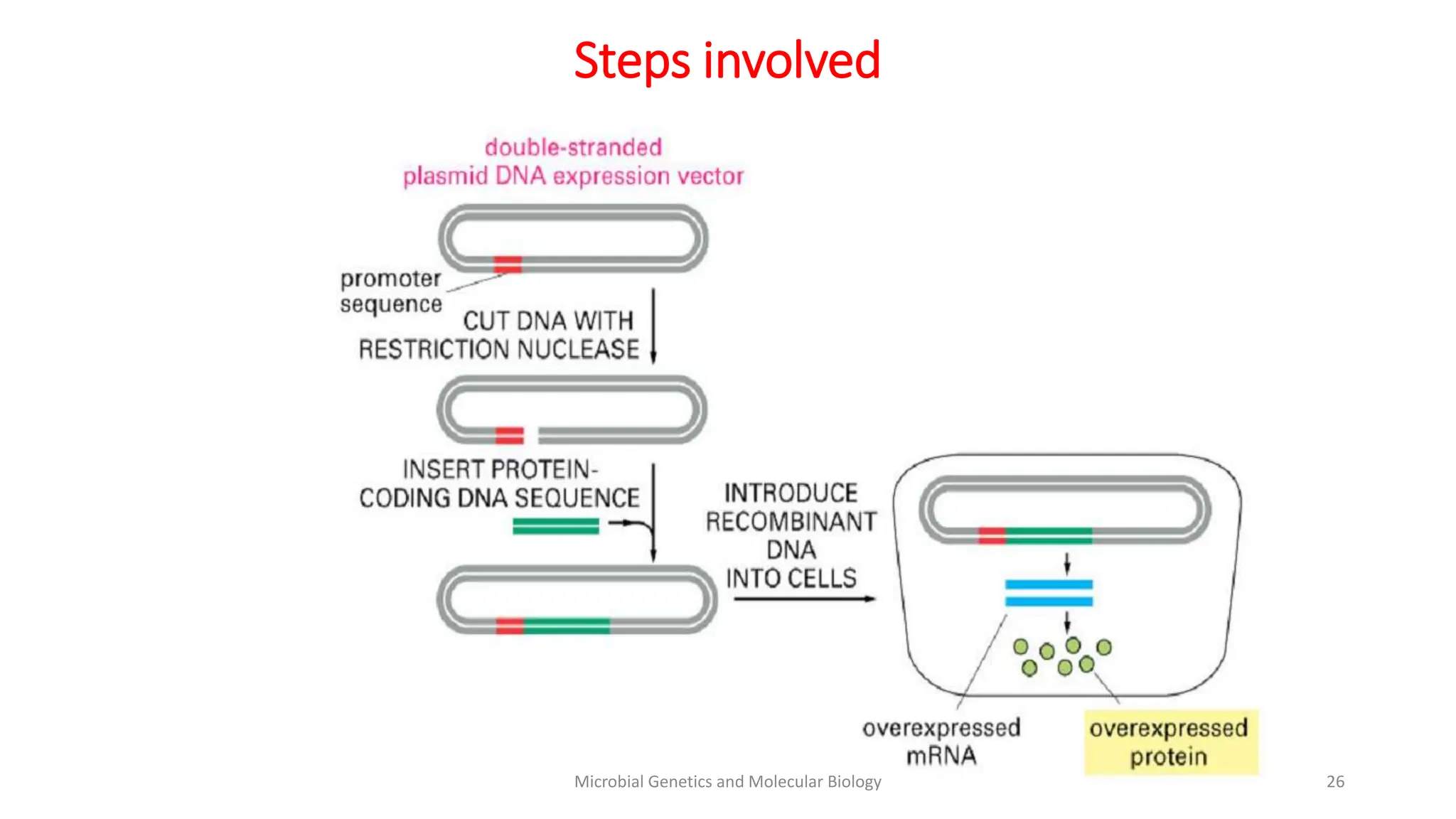
Strain improvement involves manipulating microbial strains to enhance their metabolic capacities for biotechnology applications. Targets of strain improvement include rapid growth, genetic stability, non-toxicity, large cell size, ability to use cheaper substrates, increased productivity, and reduced cultivation costs. Traditional methods of strain improvement include mutagenesis, classical genetics, and rational selection. Modern techniques utilize genetic engineering and recombinant DNA technology to introduce desired mutations for traits like increased thermostability or altered substrate range.