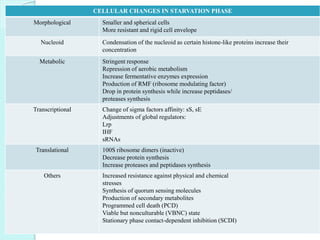
The document discusses starvation stress responses in bacteria. It describes how bacteria like E. coli and Salmonella induce universal stress proteins and alter gene expression through mechanisms like the stringent response when nutrients become scarce. This prepares the bacterial cells to survive extended periods of starvation by stopping growth, adapting their metabolism, and developing resistant cell morphologies. Key aspects of the starvation stress response discussed include the roles of RpoS, ppGpp, and morphological changes that increase resistance of bacterial cells to stress.