This document discusses ways to make R code more reproducible through cleaner coding practices, functional programming, and collaboration tools. It recommends:
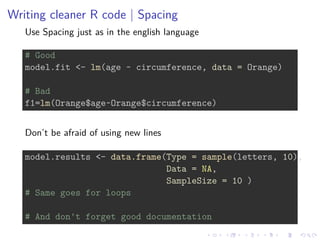
1. Writing cleaner code through descriptive names, spacing, and documentation.
2. Functionalizing code by defining reusable functions to avoid repetition and improve flexibility.
3. Using pipes to chain together functions and solve complex problems through simple pieces.
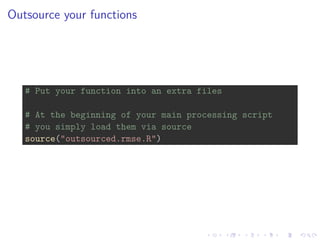
4. Outsourcing functions to external files and version controlling code with Git and GitHub to enable collaboration.





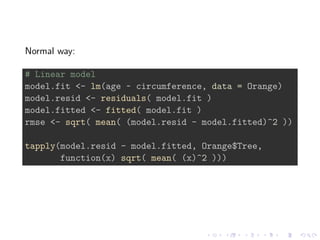

![model.fit <- lm(age ~ circumference, data = Orange)
# This function is more flexible, can be further customized
# applied in other situations
rmse(model.fit)
## [1] 1041.809
rmse(model.fit, Orange$Tree)
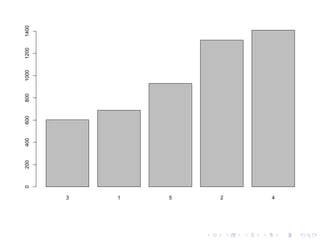
## 3 1 5 2 4
## 602.4244 688.8896 929.9055 1319.1573 1408.7033](https://image.slidesharecdn.com/reproducibility-150325044245-conversion-gate01/85/Reproducibility-with-R-12-320.jpg)

![library(dplyr)
model.rmse <- Orange %>%
lm(age ~ circumference, data=.) %>%
rmse(., Orange$Tree) %>%
barplot
OR like this (Correlation within Iris dataset)
iris %>% group_by(Species) %>%
summarize(count = n(), pear_r = cor(Sepal.Length, Petal.L
arrange(desc(pear_r))
## Source: local data frame [3 x 3]
##
## Species count pear_r
## 1 virginica 50 0.8642247
## 2 versicolor 50 0.7540490
## 3 setosa 50 0.2671758](https://image.slidesharecdn.com/reproducibility-150325044245-conversion-gate01/85/Reproducibility-with-R-14-320.jpg)