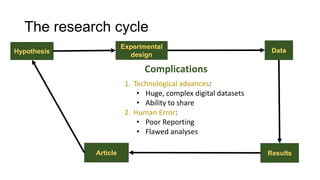
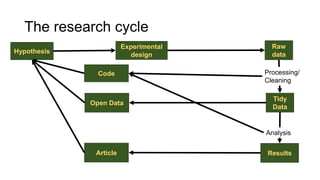
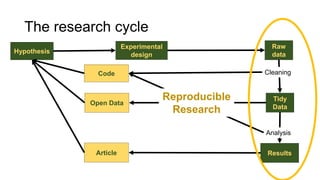
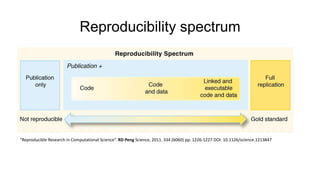
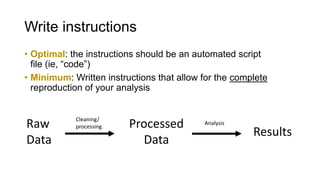
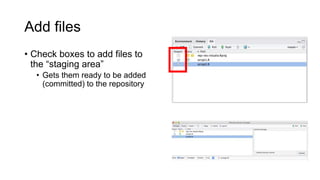
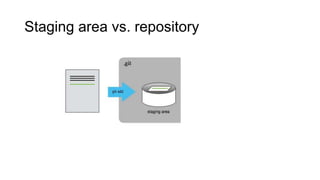
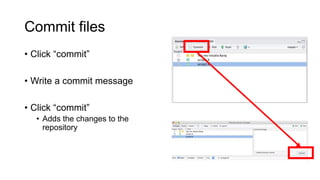
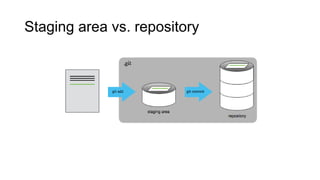
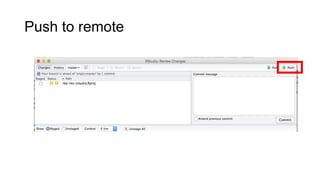
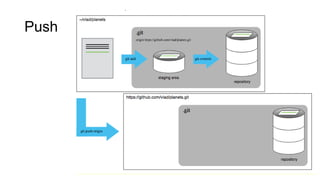
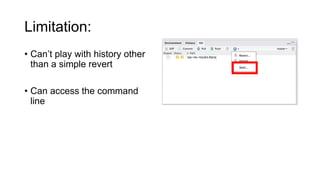
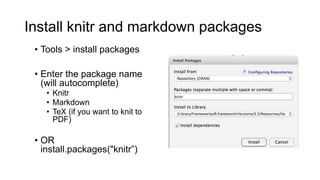
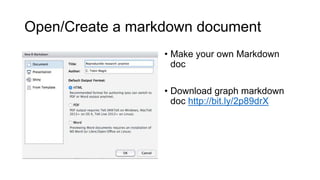
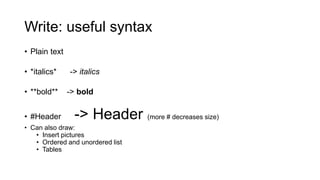
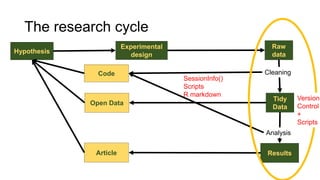
This document discusses reproducible research and provides guidance on how to conduct analysis in a reproducible manner. Reproducible research means distributing all data, code, and tools required to reproduce published results. Key aspects include automating analysis, using version control like Git, and producing human and machine readable reports in R Markdown. The presenter provides examples of documenting analysis in R and using R Markdown documents. Researchers are encouraged to think about reproducibility in their entire workflow and use checklists to ensure all elements like data, code, software details are preserved.