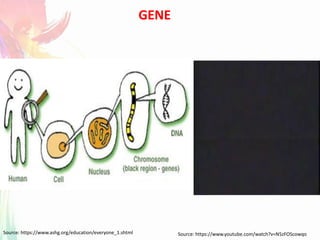
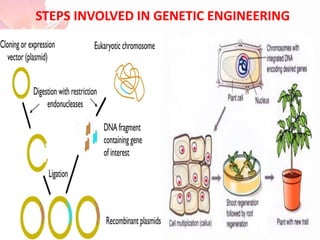
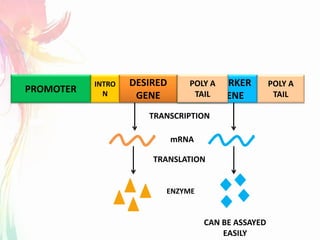
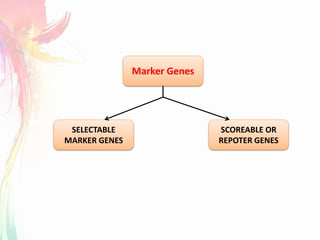
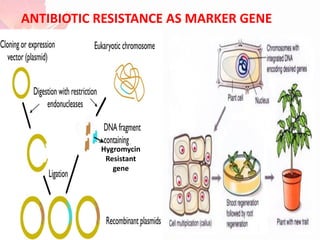
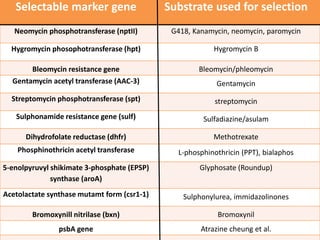
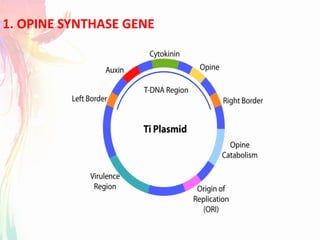
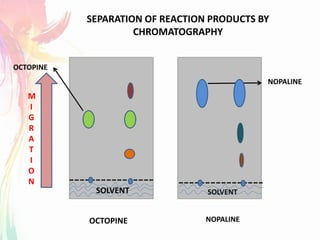
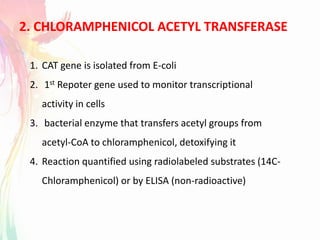
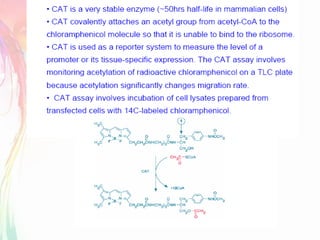
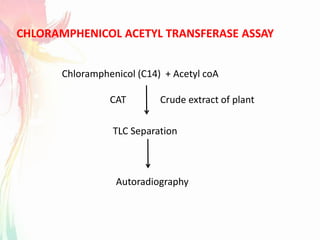
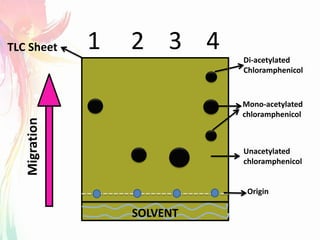
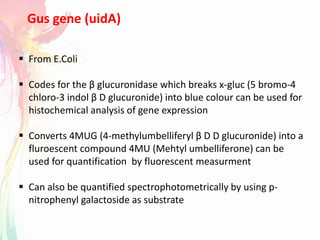
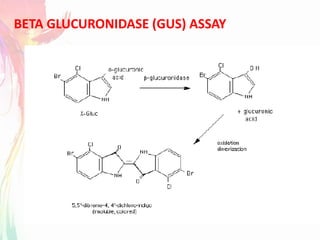
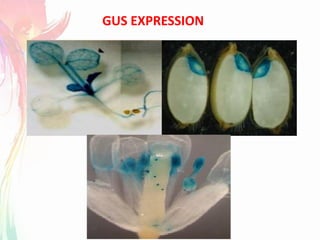
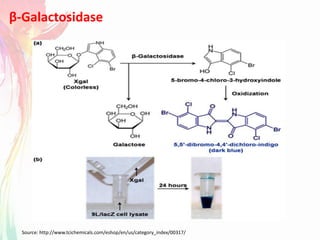
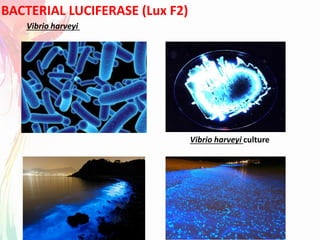
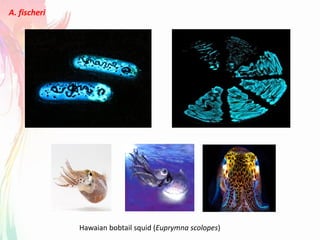
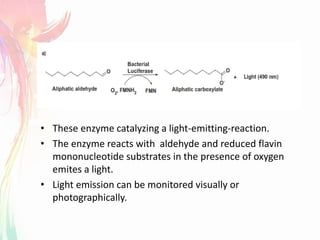
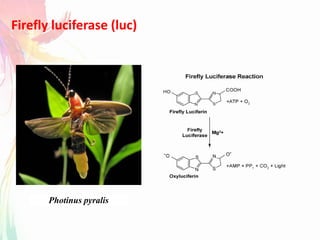
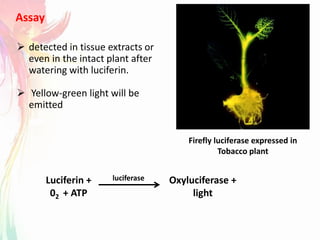
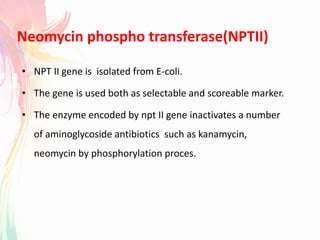
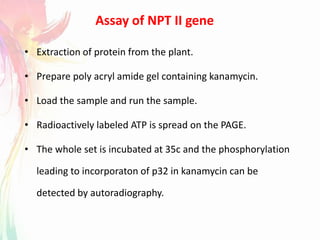
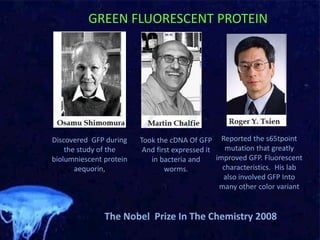
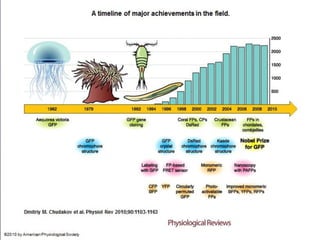
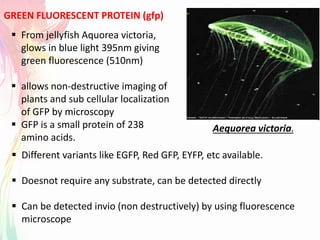
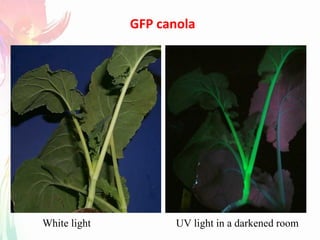
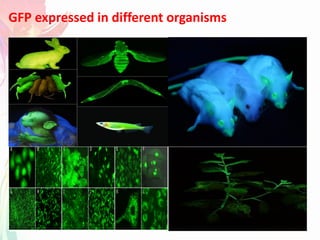
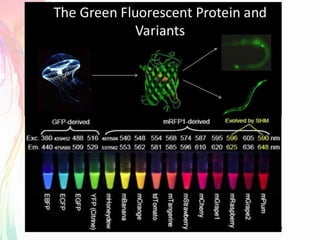
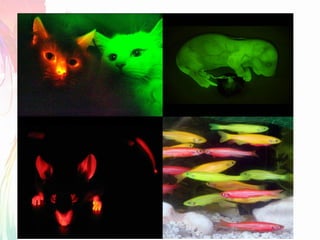
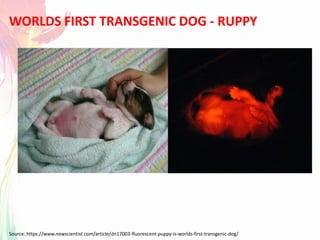
The document discusses the mechanisms and types of genetic engineering, highlighting the use of selectable and reporter genes such as hygromycin resistant gene, beta-glucuronidase (GUS), and green fluorescent protein (GFP). It details the steps involved in assaying these genes and their importance in plant biotechnology, including various methods for detecting gene expression. The document also mentions the discovery of GFP and its applications, culminating in the creation of the world's first transgenic dog.