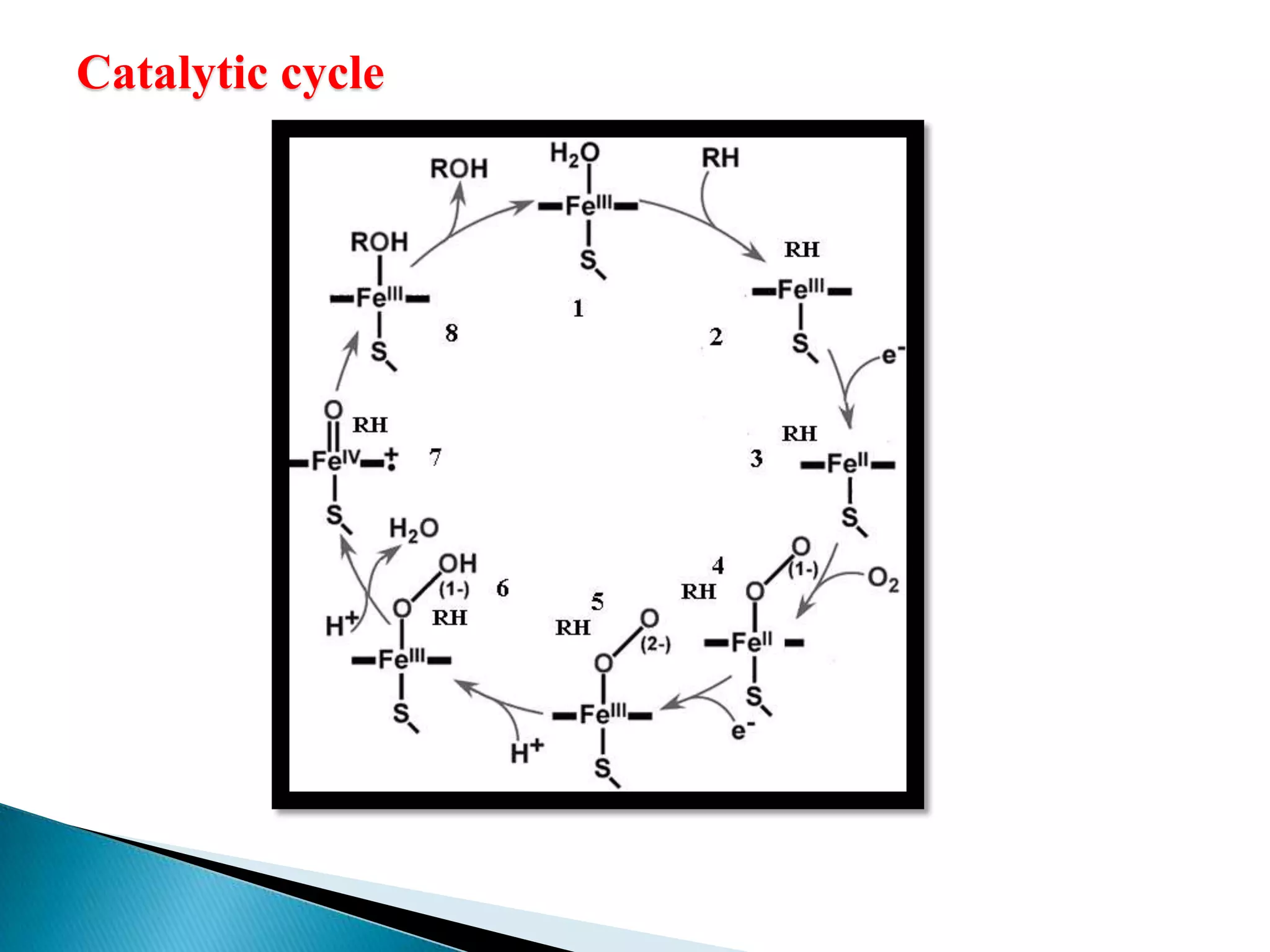
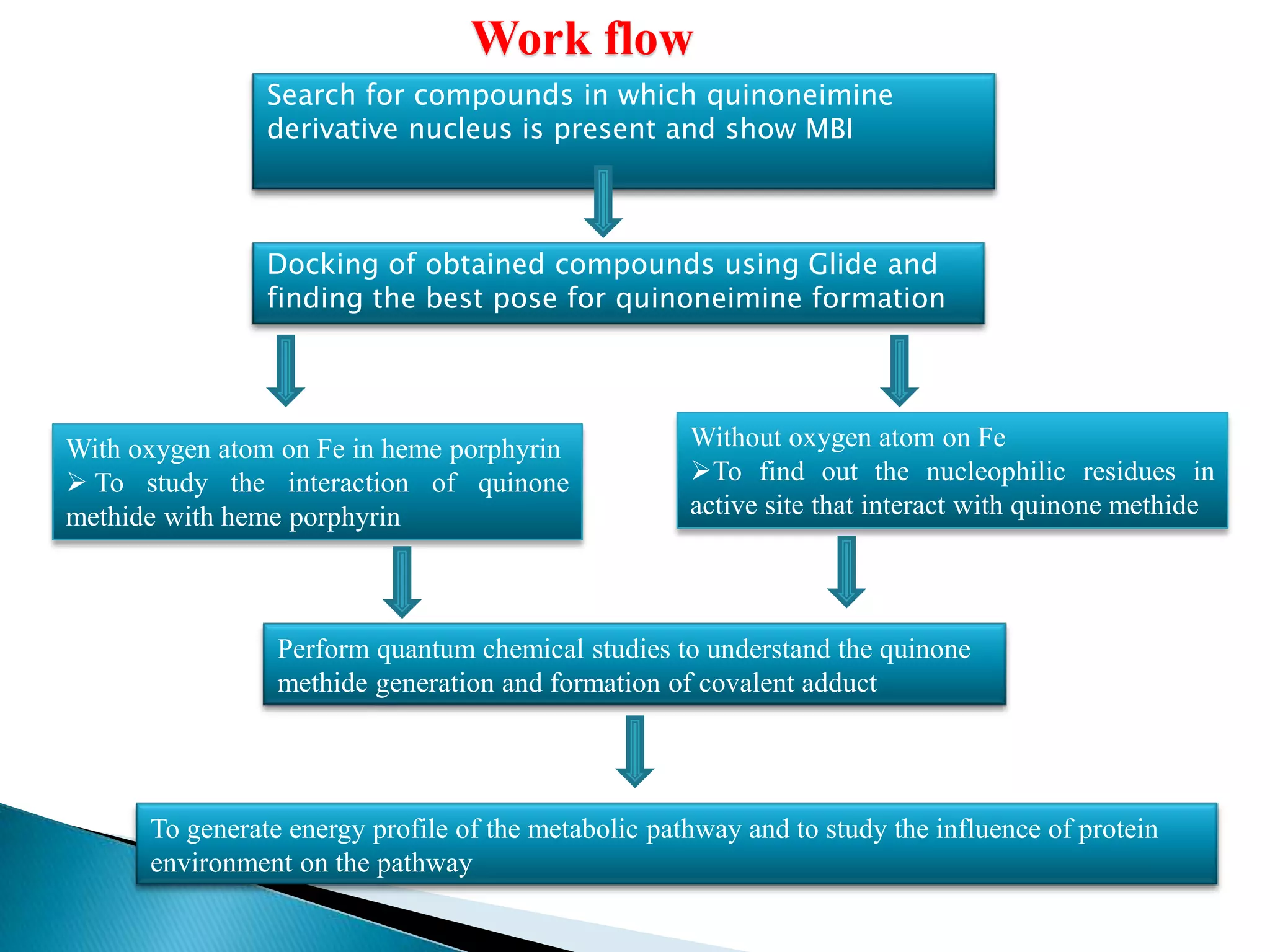
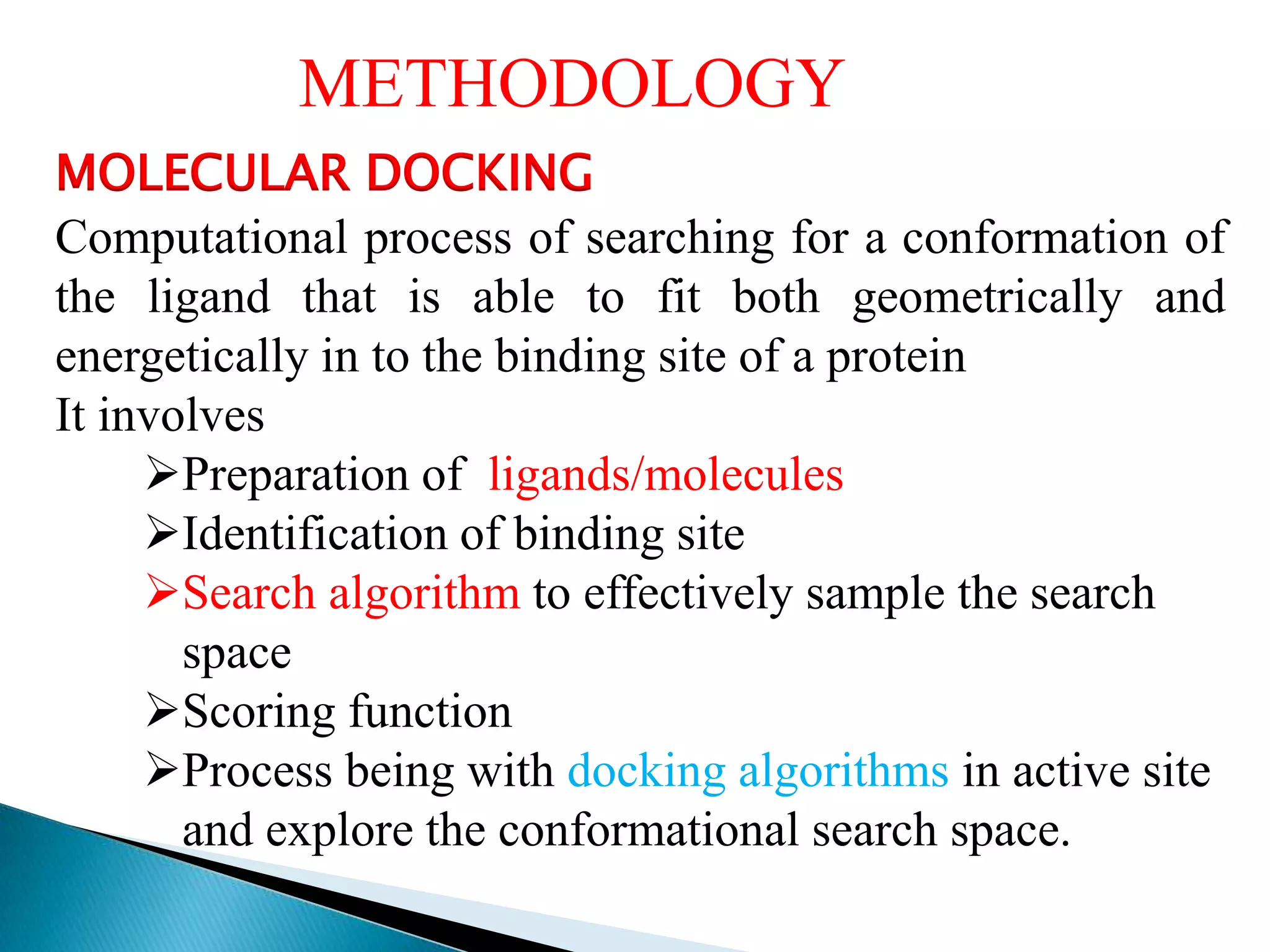
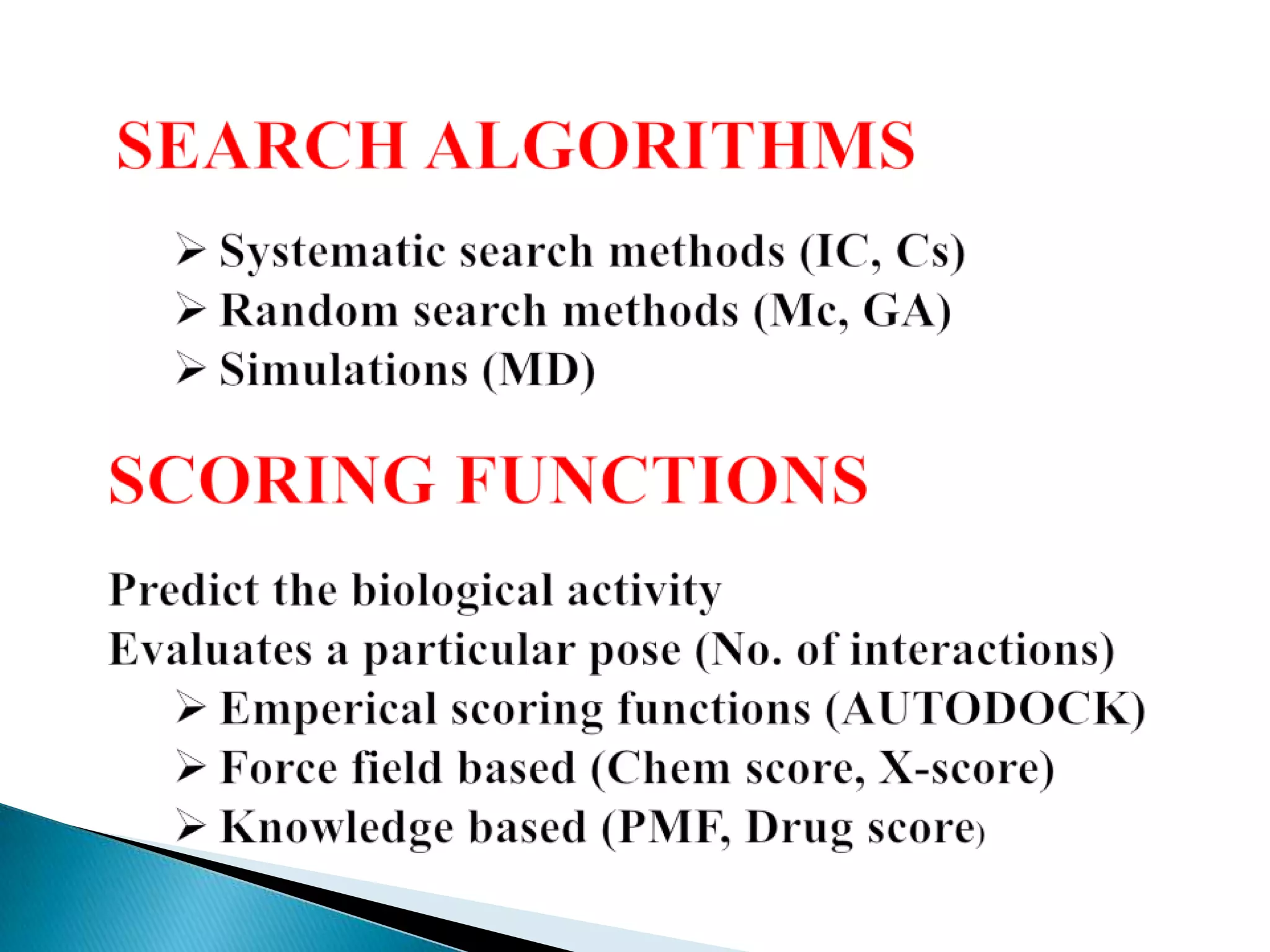
The document outlines a presentation on drug metabolism, focusing on the role of cytochrome P450 enzymes in the biochemical modification of drugs. It discusses various phases of drug reactions, including functionalization and conjugation, along with the mechanisms of enzyme inhibition and the significance of molecular interactions. The research objectives include studying the formation of quinone methide metabolites and understanding their impact on cytochrome P450s using quantum chemical methods and molecular docking analyses.