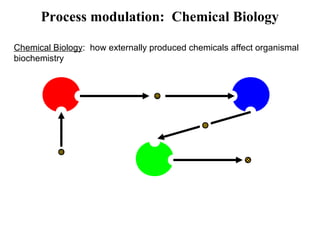
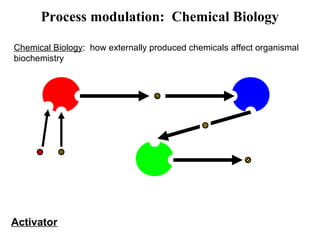
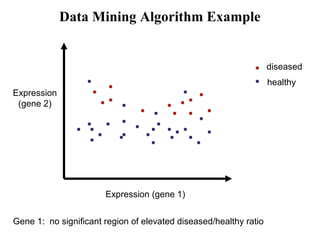
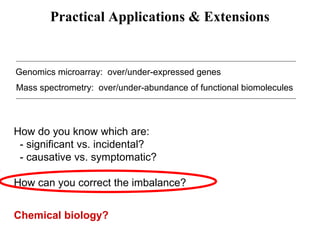
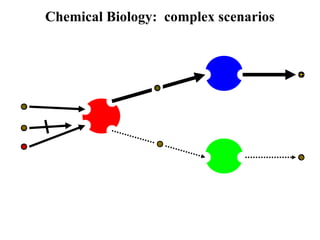
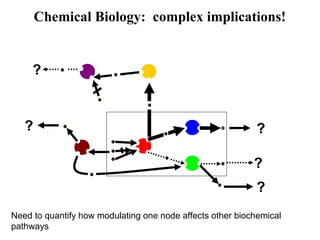
This document discusses approaches for personalized medicine through molecular interrogation, data mining, and systems biology. It describes how molecular interrogation techniques like microarrays and mass spectrometry can be used to understand biochemical processes in the body and identify biomarkers for diseases. Data mining algorithms can then determine which gene expression and molecular abundance anomalies are significant and causative for specific diseases. Systems biology modeling examines how modulating particular biochemical processes affects organism-wide pathways and can help predict patient responses, side effects, and personalized treatment options through approaches like varying drug doses or combinations.
















![a
b
x
c
d
e
f
A
B
C
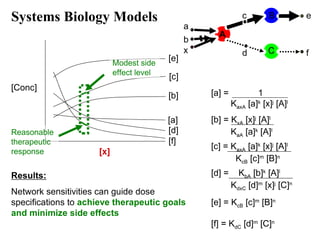
[c] = KaxA [a]k
[x]j
[A]l
KcB [c]m
[B]n
[d] = KbA [b]k
[A]l
KdxC [d]m
[x]j
[C]n
[e] = KcB [c]m
[B]n
[f] = KdC [d]m
[C]n
[a] = 1
KaxA [a]k
[x]j
[A]l
[b] = KxA [x]j
[A]l
KaA [a]k
[A]l
Systems Biology Models
[Conc]
time
[a]
[d]
[f]
[c]
[b]
[e]
x administered
Procedure:
Microarray, MS or chemical biology data
Record multiple time points
Perturb the system (i.e., add x)
Fit concentrations to coupled equations](https://image.slidesharecdn.com/personalizedmedicine-130123101450-phpapp01-140406132509-phpapp01/85/Personalized-medicine-29-320.jpg)
![a
b
x
c
d
e
f
A
B
C
[c] = KaxA [a]k
[x]j
[A]l
KcB [c]m
[B]n
[d] = KbA [b]k
[A]l
KdxC [d]m
[x]j
[C]n
[e] = KcB [c]m
[B]n
[f] = KdC [d]m
[C]n
[a] = 1
KaxA [a]k
[x]j
[A]l
[b] = KxA [x]j
[A]l
KaA [a]k
[A]l
Systems Biology Models
[Conc]
time
[a]
[d]
[f]
[c]
[b]
[e]
x administered
Results:
Network sensitivities can pinpoint possible side
effects](https://image.slidesharecdn.com/personalizedmedicine-130123101450-phpapp01-140406132509-phpapp01/85/Personalized-medicine-30-320.jpg)
![a
b
x
c
d
e
f
A
B
C
[c] = KaxA [a]k
[x]j
[A]l
KcB [c]m
[B]n
[d] = KbA [b]k
[A]l
KdxC [d]m
[x]j
[C]n
[e] = KcB [c]m
[B]n
[f] = KdC [d]m
[C]n
[a] = 1
KaxA [a]k
[x]j
[A]l
[b] = KxA [x]j
[A]l
KaA [a]k
[A]l
Systems Biology Models
[Conc]
time
[a]
[d]
[f]
[c]
[b]
[e]
x administered
Procedure:
Examine difference patient responses](https://image.slidesharecdn.com/personalizedmedicine-130123101450-phpapp01-140406132509-phpapp01/85/Personalized-medicine-31-320.jpg)
![a
b
x
c
d
e
f
A
B
C
[c] = KaxA [a]k
[x]j
[A]l
KcB [c]m
[B]n
[d] = KbA [b]k
[A]l
KdxC [d]m
[x]j
[C]n
[e] = KcB [c]m
[B]n
[f] = KdC [d]m
[C]n
[a] = 1
KaxA [a]k
[x]j
[A]l
[b] = KxA [x]j
[A]l
KaA [a]k
[A]l
Systems Biology Models
Results:
Patient 2 has decreased susceptibility to side
effects
May be able to boost dosage
without negative consequences
[Conc]
time
[a]
[d]
[f]
[c]
[b]
[e]
x administered](https://image.slidesharecdn.com/personalizedmedicine-130123101450-phpapp01-140406132509-phpapp01/85/Personalized-medicine-32-320.jpg)
![a
b
x
c
d
e
f
A
B
C
[c] = KaxA [a]k
[x]j
[A]l
KcB [c]m
[B]n
[d] = KbA [b]k
[A]l
KdxC [d]m
[x]j
[C]n
[e] = KcB [c]m
[B]n
[f] = KdC [d]m
[C]n
[a] = 1
KaxA [a]k
[x]j
[A]l
[b] = KxA [x]j
[A]l
KaA [a]k
[A]l
Systems Biology Models
[Conc]
time
[a]
[d]
[f]
[c]
[b]
[e]
x administered
Results:
Patient 3 has diminished therapeutic
response
May need to find another drug
or target or also address [c]](https://image.slidesharecdn.com/personalizedmedicine-130123101450-phpapp01-140406132509-phpapp01/85/Personalized-medicine-33-320.jpg)
![a
b
x
c
d
e
f
A
B
C
[c] = KaxA [a]k
[x]j
[A]l
KcB [c]m
[B]n
[d] = KbA [b]k
[A]l
KdxC [d]m
[x]j
[C]n
[e] = KcB [c]m
[B]n
[f] = KdC [d]m
[C]n
[a] = 1
KaxA [a]k
[x]j
[A]l
[b] = KxA [x]j
[A]l
KaA [a]k
[A]l
Systems Biology Models
[Conc]
[x]
[d]
[f]
[a]
[b]
[c]
[e]
Procedure:
Microarray, MS or chemical biology data
Record multiple dose response points
Time average
Fit concentrations to coupled equations](https://image.slidesharecdn.com/personalizedmedicine-130123101450-phpapp01-140406132509-phpapp01/85/Personalized-medicine-34-320.jpg)