The document discusses an RNA-seq workshop agenda with the following key points:
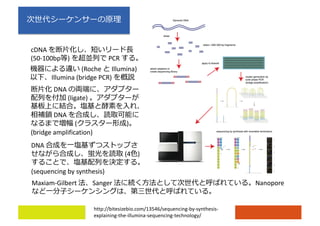
1) It includes sessions on RNA-seq introduction, analysis workflows from mapping to differential expression and enrichment analysis, and available online resources and tools for RNA-seq data analysis.
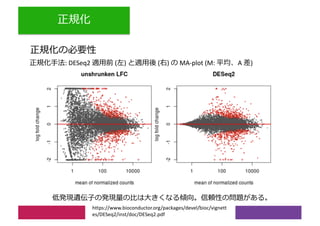
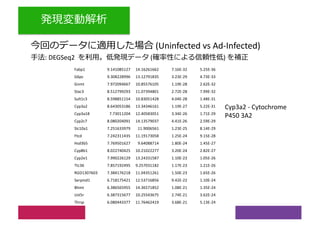
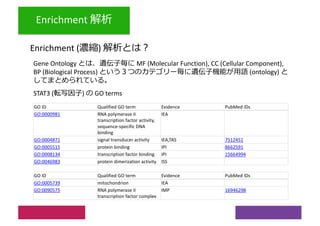
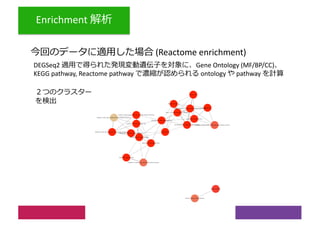
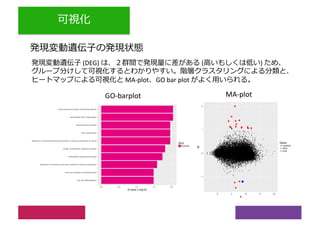
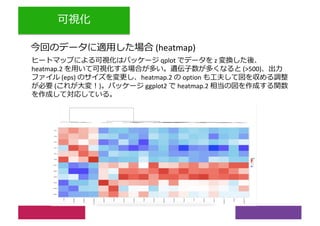
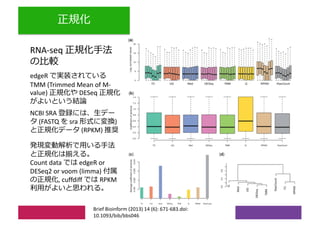
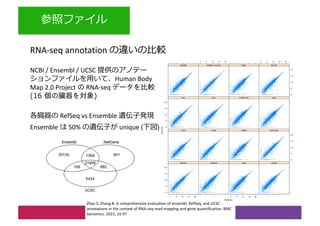
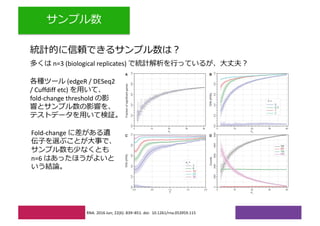
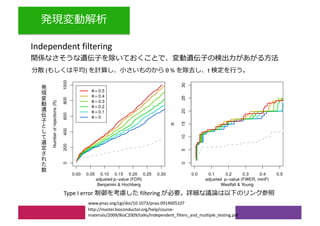
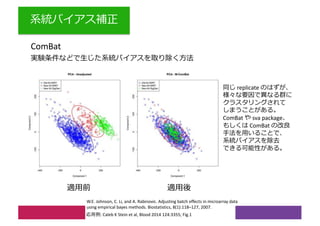
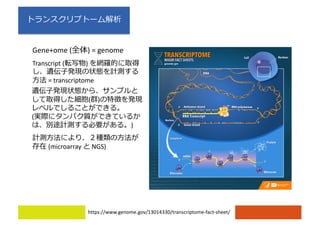
2) Differential expression analysis will be performed using DESeq2 and genes will be analyzed for enrichment in Gene Ontology terms and KEGG pathways to identify biological processes and pathways impacted.
3) The workshop aims to introduce common RNA-seq analysis concepts and provide hands-on experience with tools like DESeq2, GO and KEGG enrichment to find differentially expressed genes and interpret results biologically.

![(NGS )
[MA/NGS]
Enrichment (GO, pathway), (heatmap, etc)
(Differential Expressed Genes: DEG) [MA/NGS]](https://image.slidesharecdn.com/rnaseqtutorial-180730090809/85/RNA-seq-tutorial-2-320.jpg)
![(NGS )
[MA/NGS]
Enrichment (GO, pathway), (heatmap, etc)
(Differential Expressed Genes: DEG) [MA/NGS]](https://image.slidesharecdn.com/rnaseqtutorial-180730090809/85/RNA-seq-tutorial-3-320.jpg)
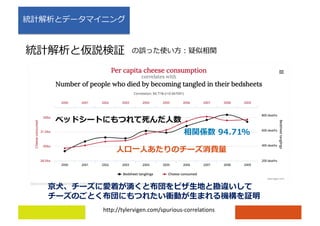
![)
(
][
[
https://www.nature.com/articles/ncomms9727](https://image.slidesharecdn.com/rnaseqtutorial-180730090809/85/RNA-seq-tutorial-7-320.jpg)

![-- [ ]
-- [ ] R and Bioconductor / command line / python
-- [ ]
-- [ ] ( )
-- [ ] (2 1 )
-- [ ] 29 NGS
https://biosciencedbc.jp/human/human-resources/workshop/h29
-- [ ] RNA-Seq ( )
-- [ ] Dr. Bono
-- [ ] ( )
script
shinzy.nakaoka@gmail.com](https://image.slidesharecdn.com/rnaseqtutorial-180730090809/85/RNA-seq-tutorial-9-320.jpg)

![(NGS )
[MA/NGS]
Enrichment (GO, pathway), (heatmap, etc)
(Differential Expressed Genes: DEG) [MA/NGS]](https://image.slidesharecdn.com/rnaseqtutorial-180730090809/85/RNA-seq-tutorial-11-320.jpg)

![Assembly, mapping
0+ 0+ + MQ F U m vt
/I BI B& - 1& - - my o bd[g - - m I Q B
DBJ IB II( x + N S FB) LE ) d . + NB A
eln b I ps m QJ A o Q NB A b o
( - 1 f f ps
- 1 BMQBJ B B A +N EFRB N m 0+ e C M AQIL
F LU E J k L DB +A - 1 f LNBCB E o [g f ps
w
) Q F U EB d x
wu r f k q o an ] n d] n
0+ - 2 R k F eln MQ F U EB NFI II F eln
q k xo an
hcf .3& e ] n
e id TT X
LLFJD y BI U](https://image.slidesharecdn.com/rnaseqtutorial-180730090809/85/RNA-seq-tutorial-15-320.jpg)