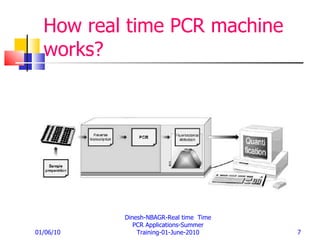
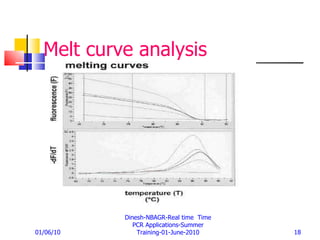
Real-time PCR provides a more accurate way to quantify gene expression compared to older methods like Northern blotting. It works by measuring fluorescence at each PCR cycle, allowing quantification of mRNA levels based on the threshold cycle. Key advantages include a wide dynamic range, high precision, and not requiring post-PCR processing. Factors like primer and probe design, normalization, and data analysis influence reproducibility and accuracy of real-time PCR gene expression analysis.