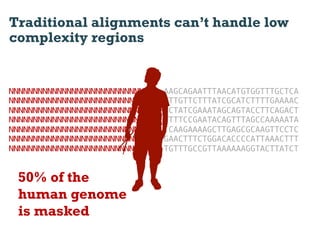
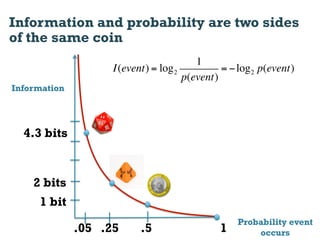
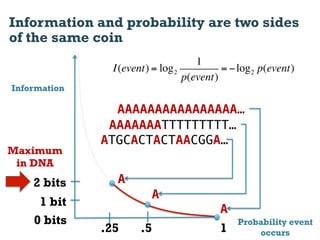
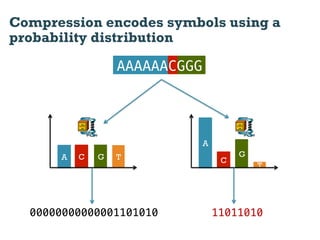
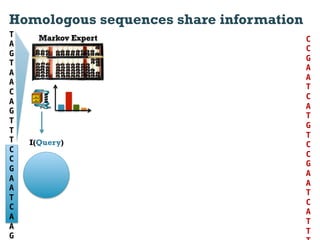
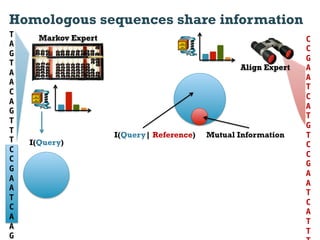
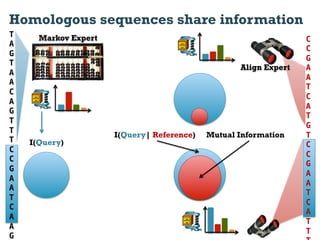
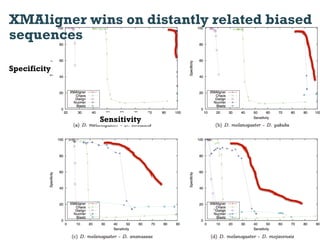
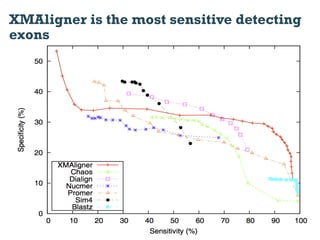
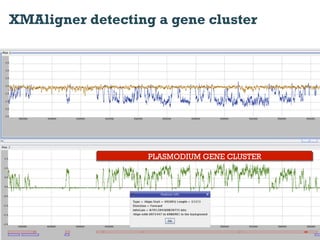
This document discusses using information compression to align biological sequences. It notes that traditional sequence alignment methods cannot handle low complexity regions or account for sequence biases. Information and probability are described as two sides of the same coin, with more similar sequences sharing more information. The document proposes aligning sequences by compressing them using a probabilistic model, asserting that homologous sequences will share information and compress better together. This alignment by compression approach is said to outperform traditional methods in aligning distantly related or biased sequences.