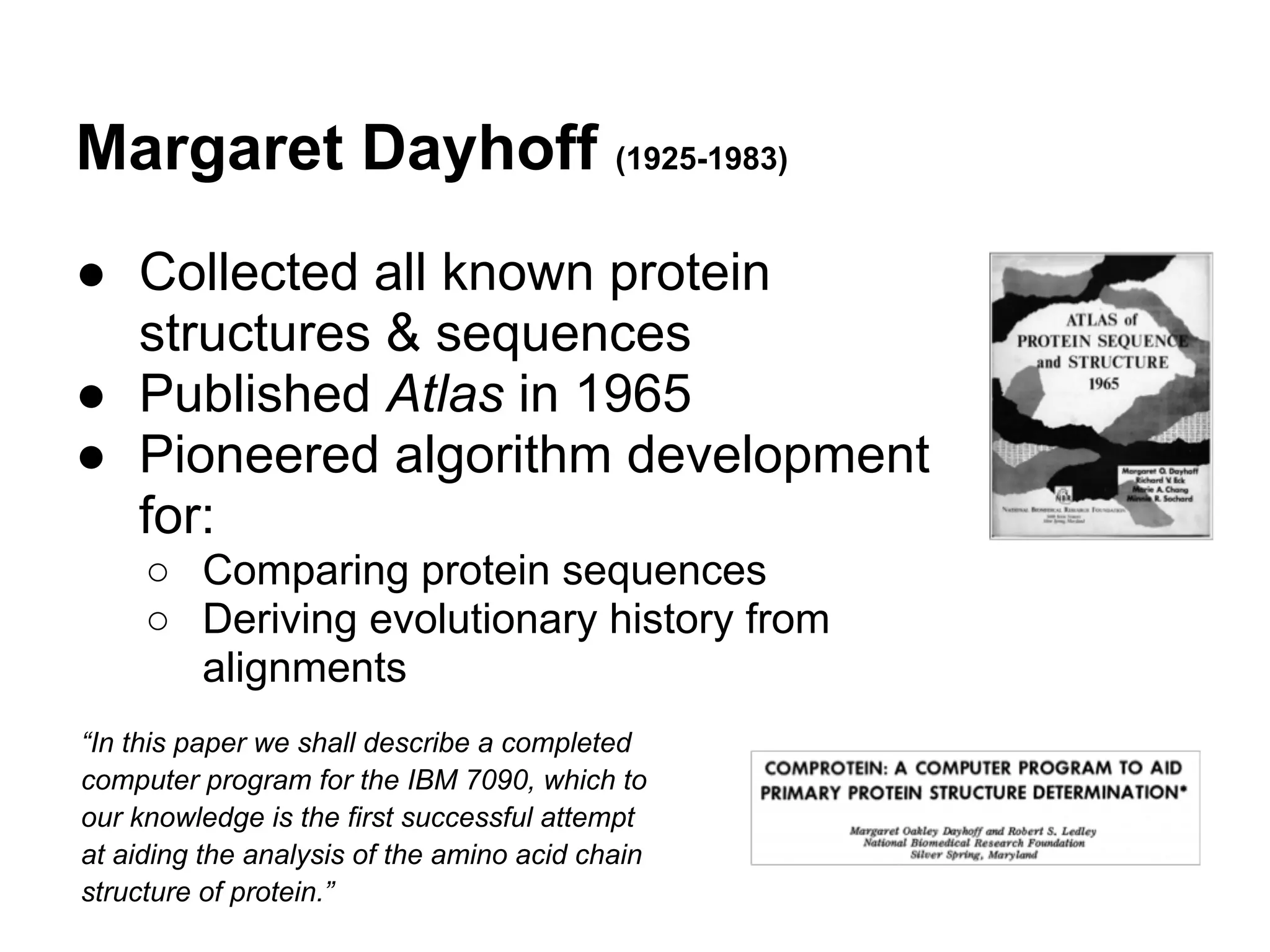
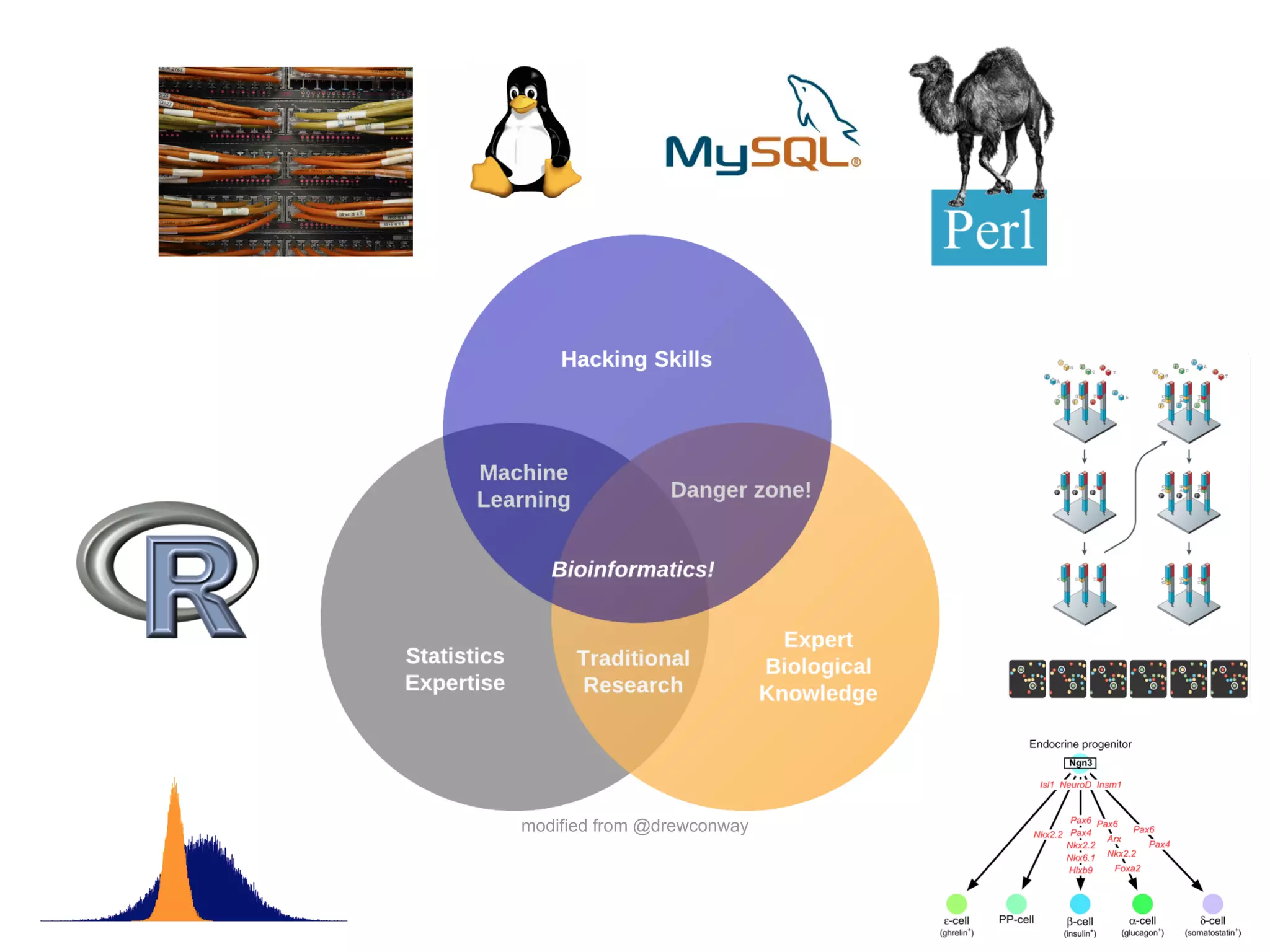
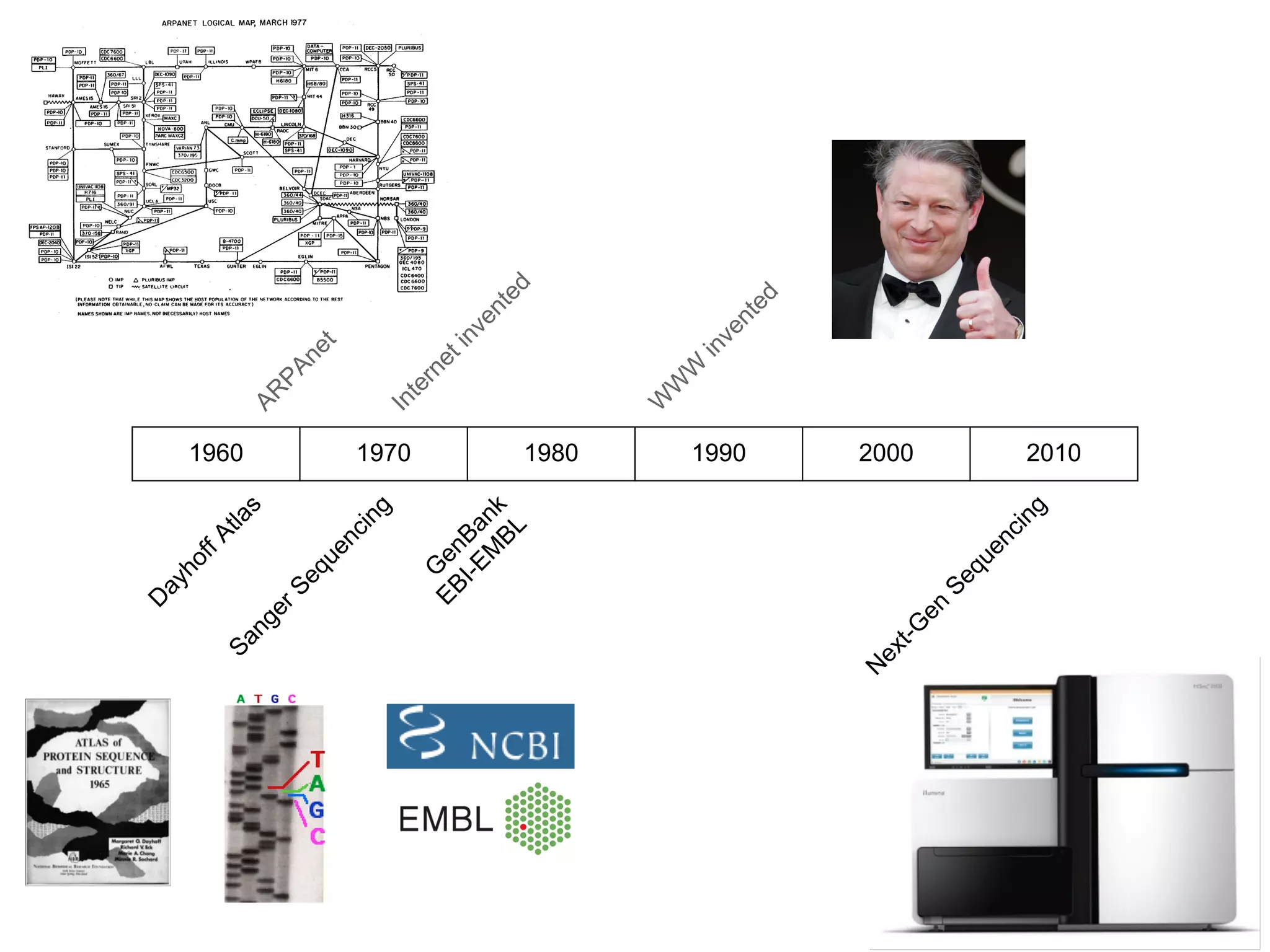
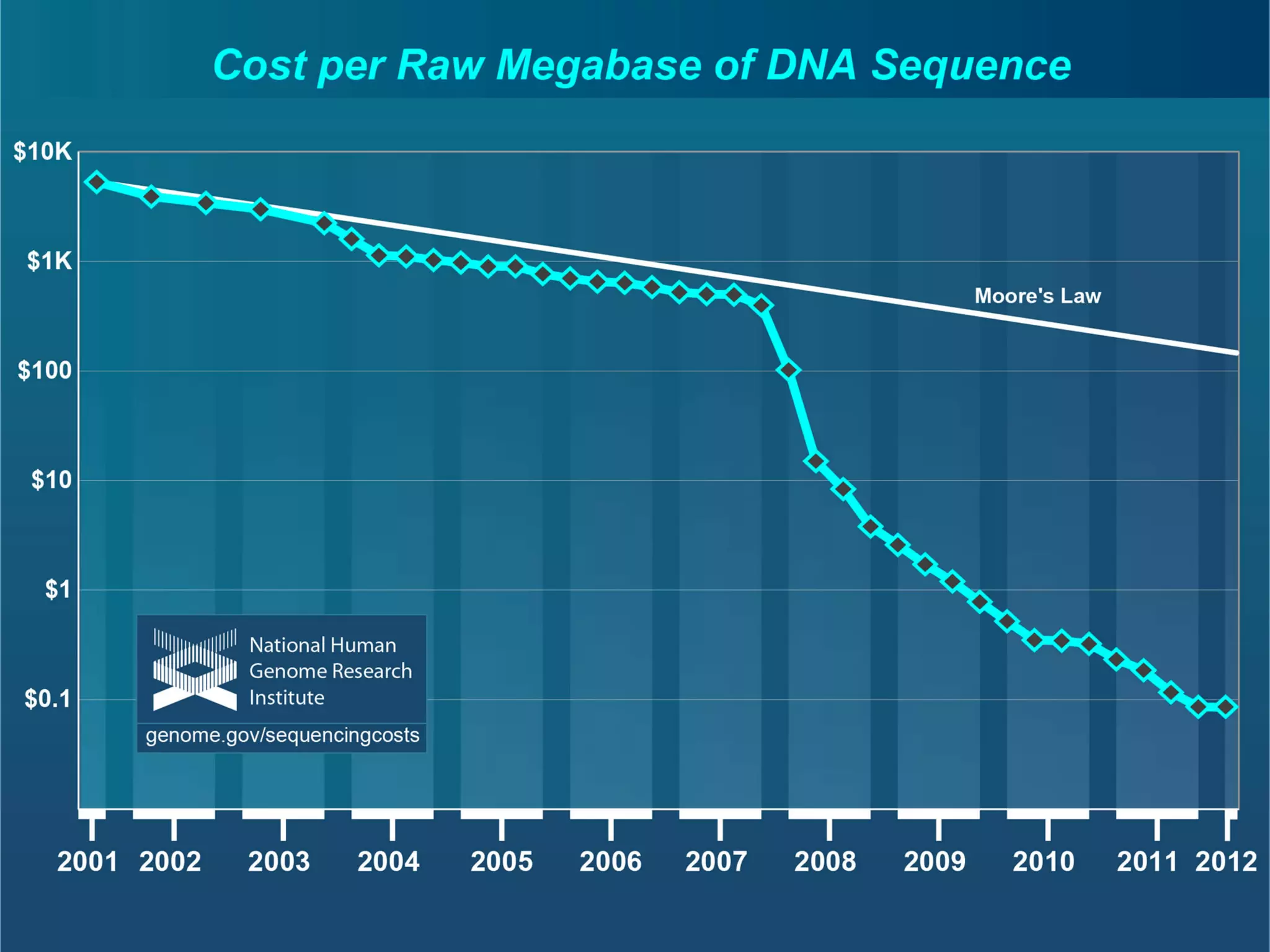
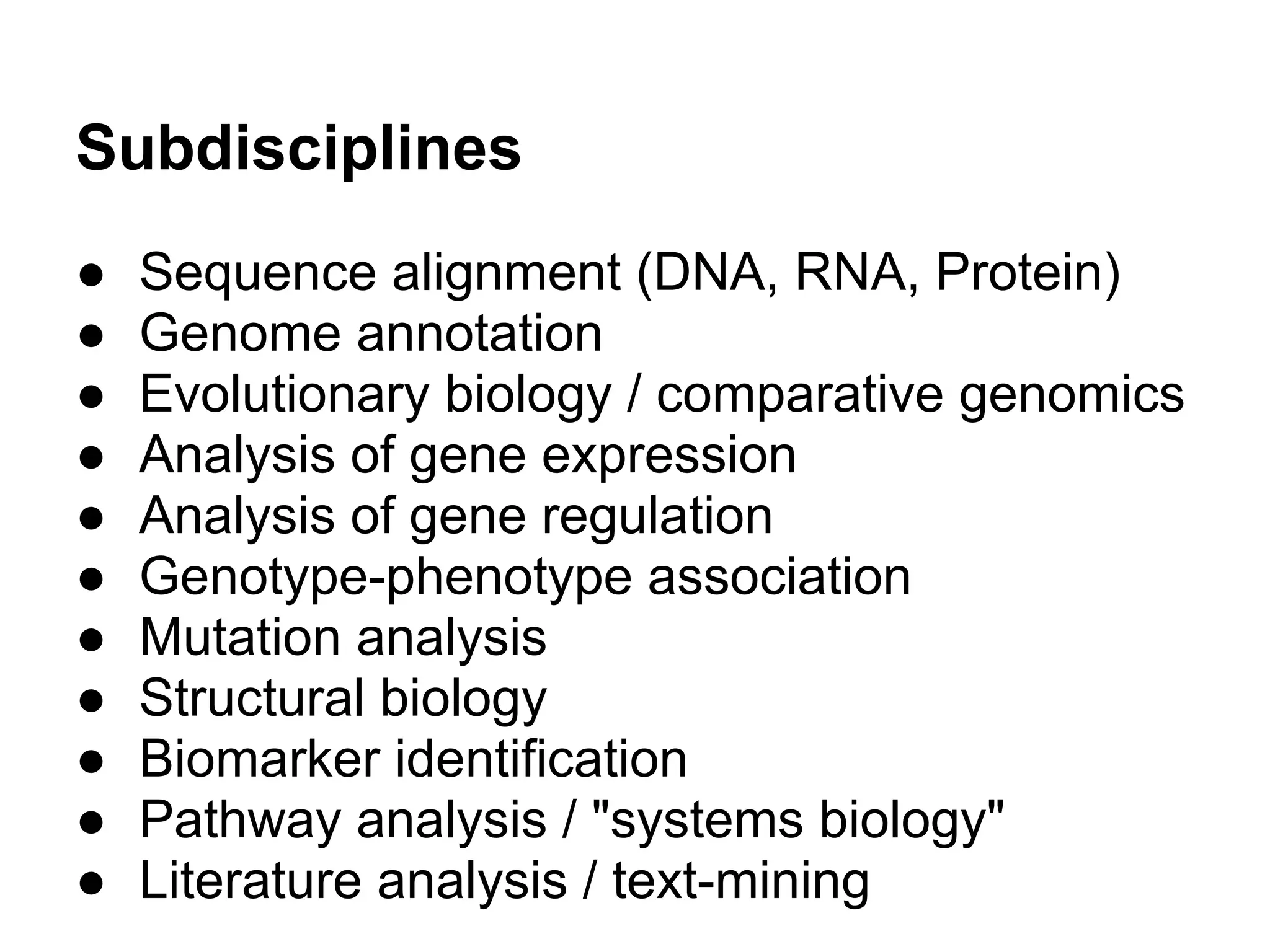
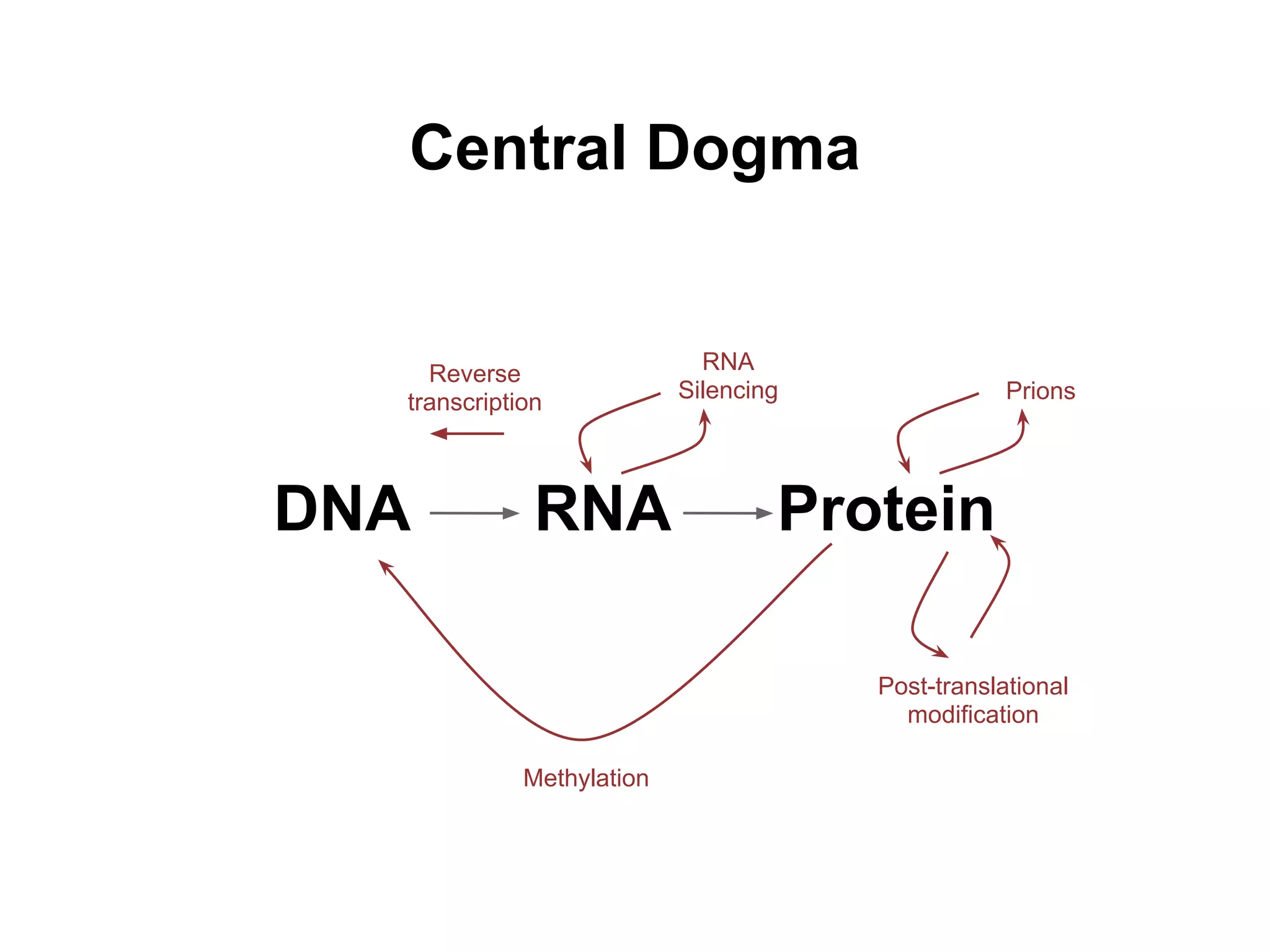
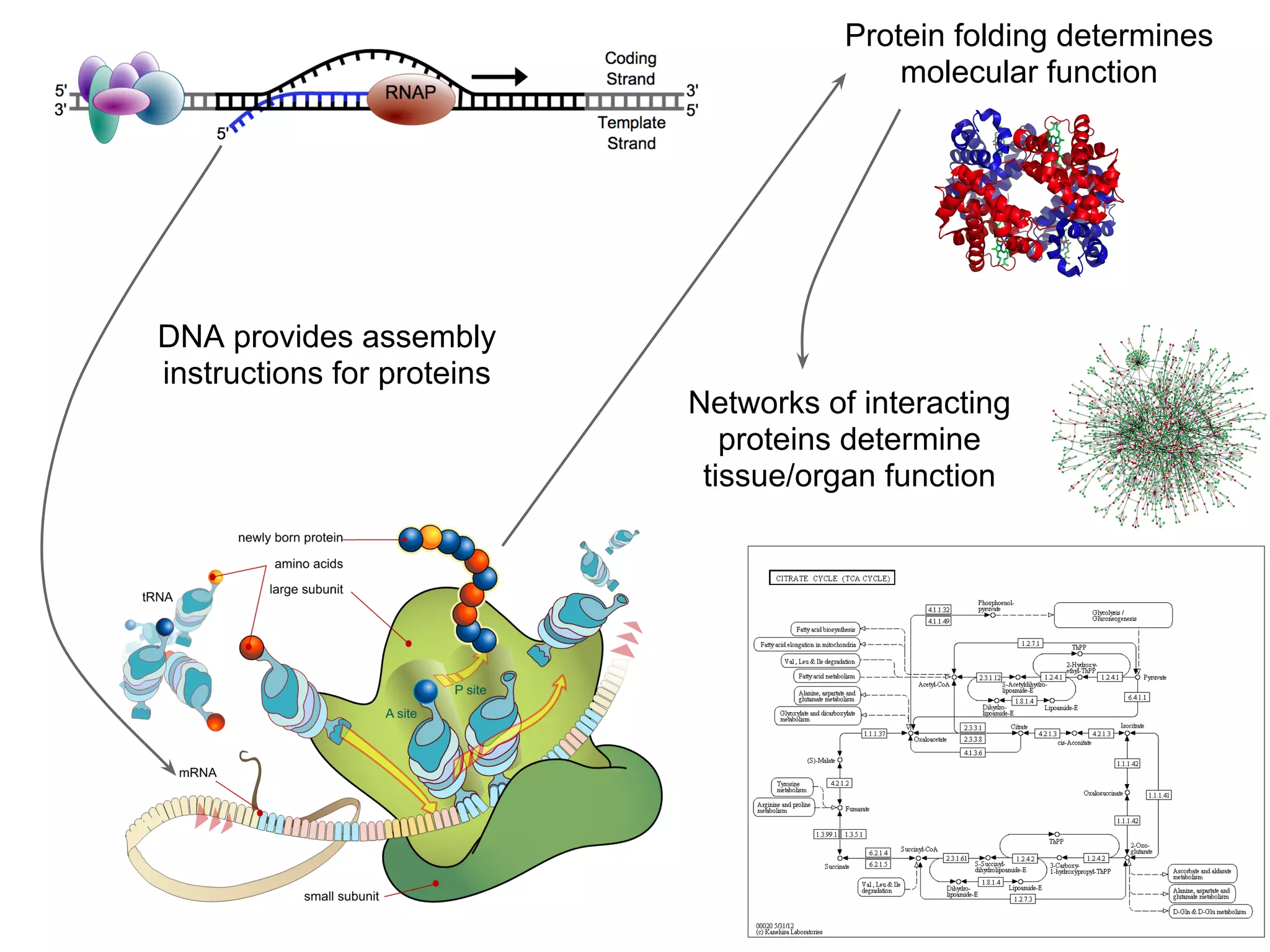
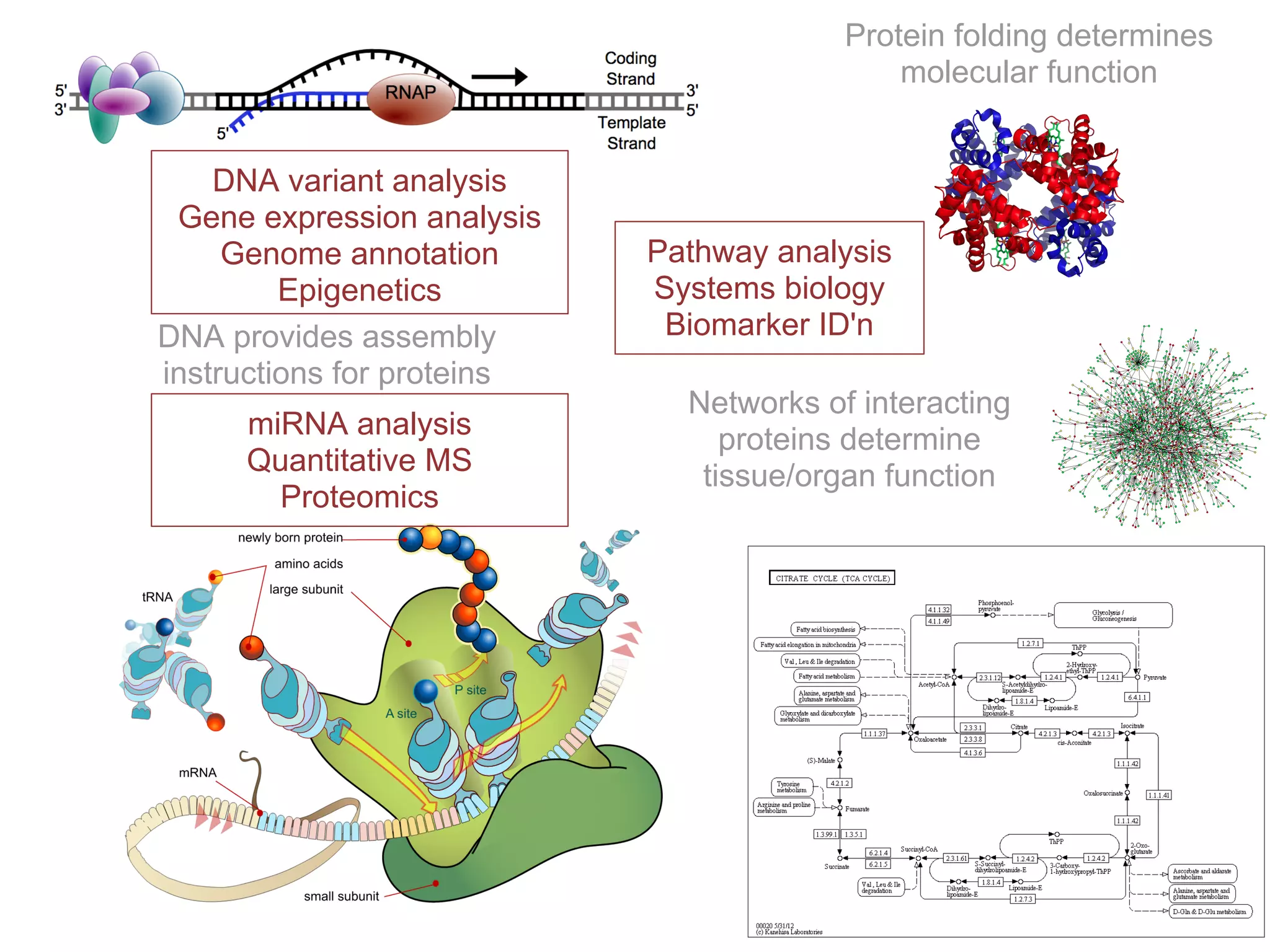
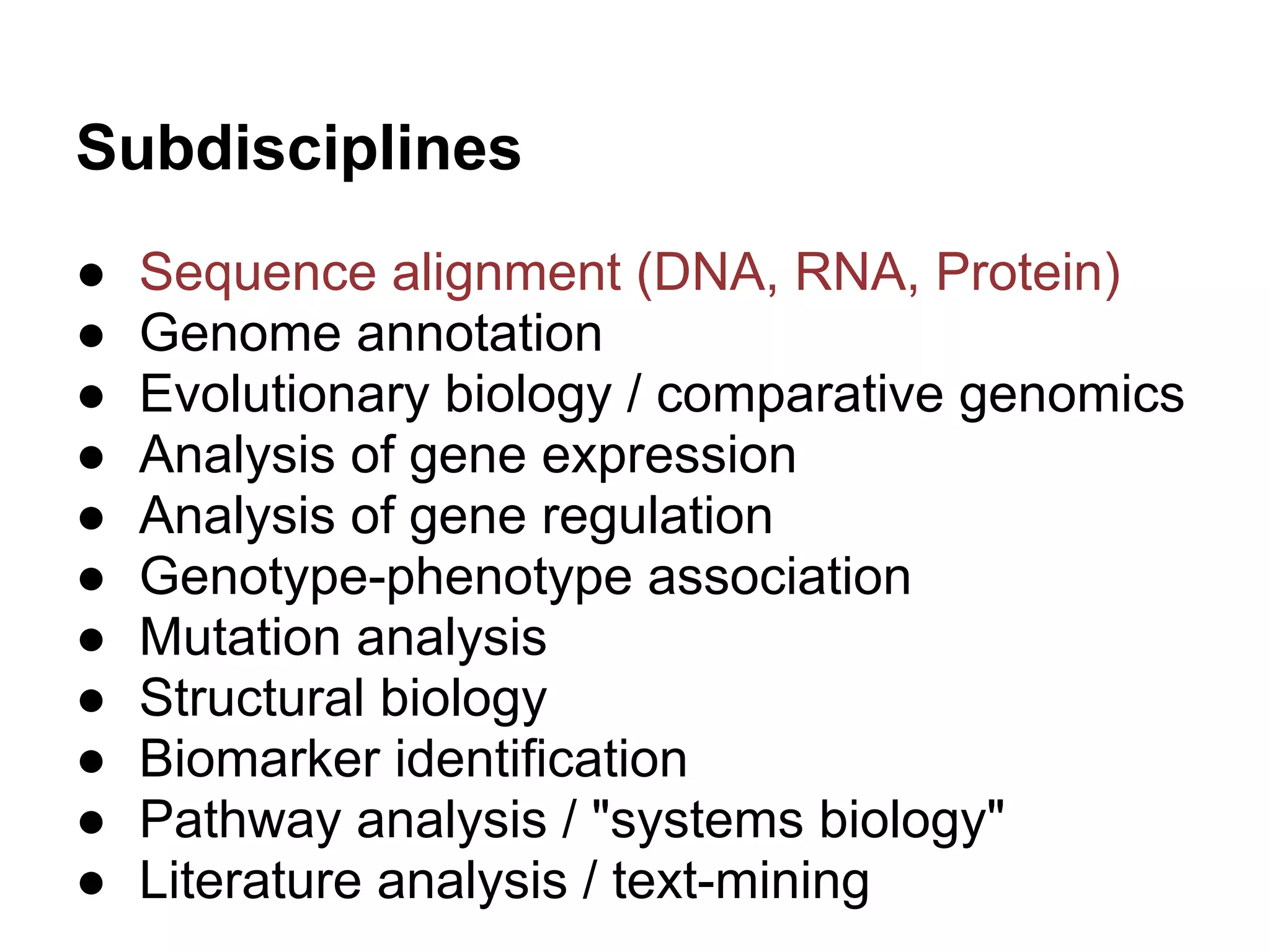
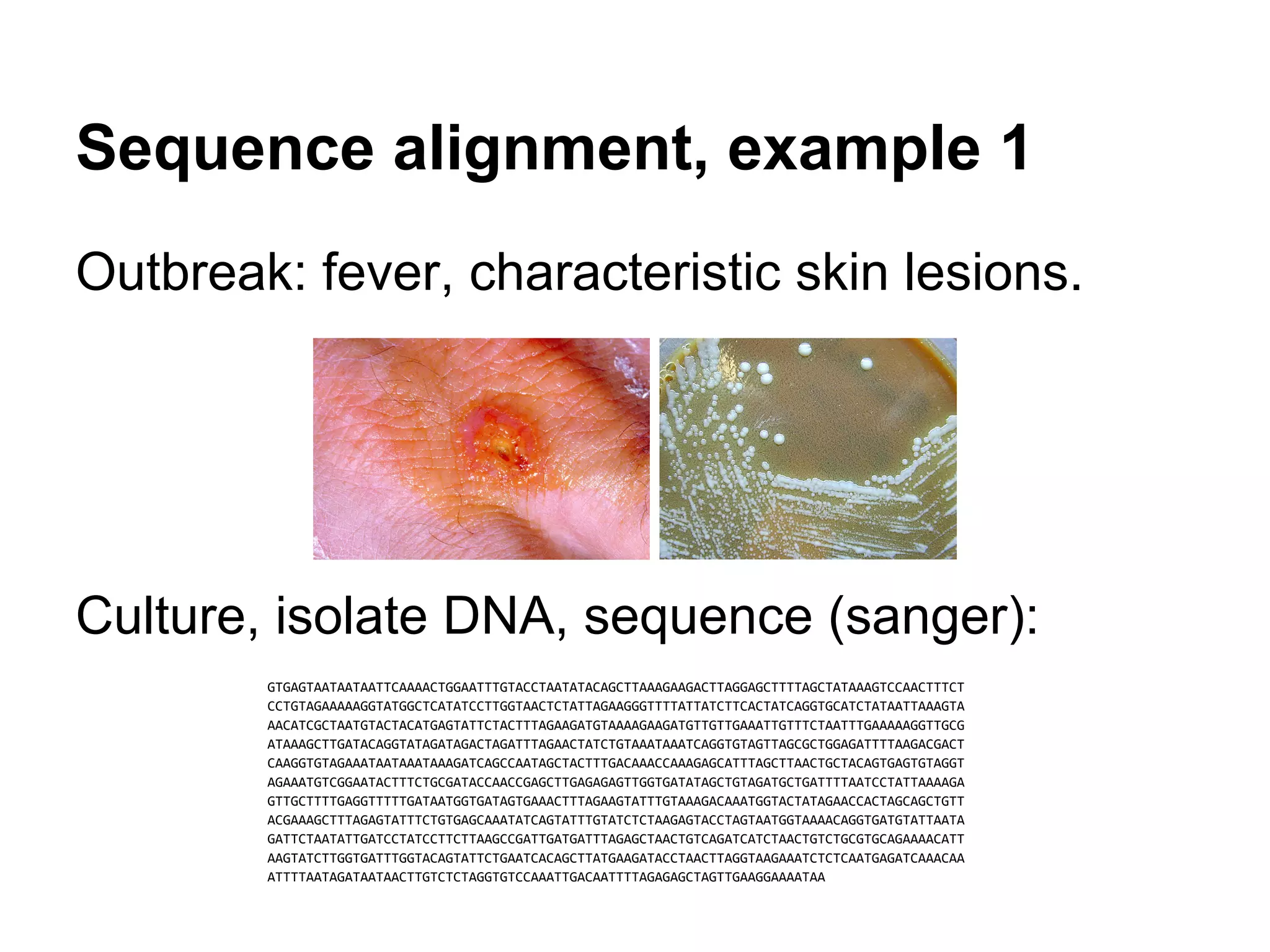
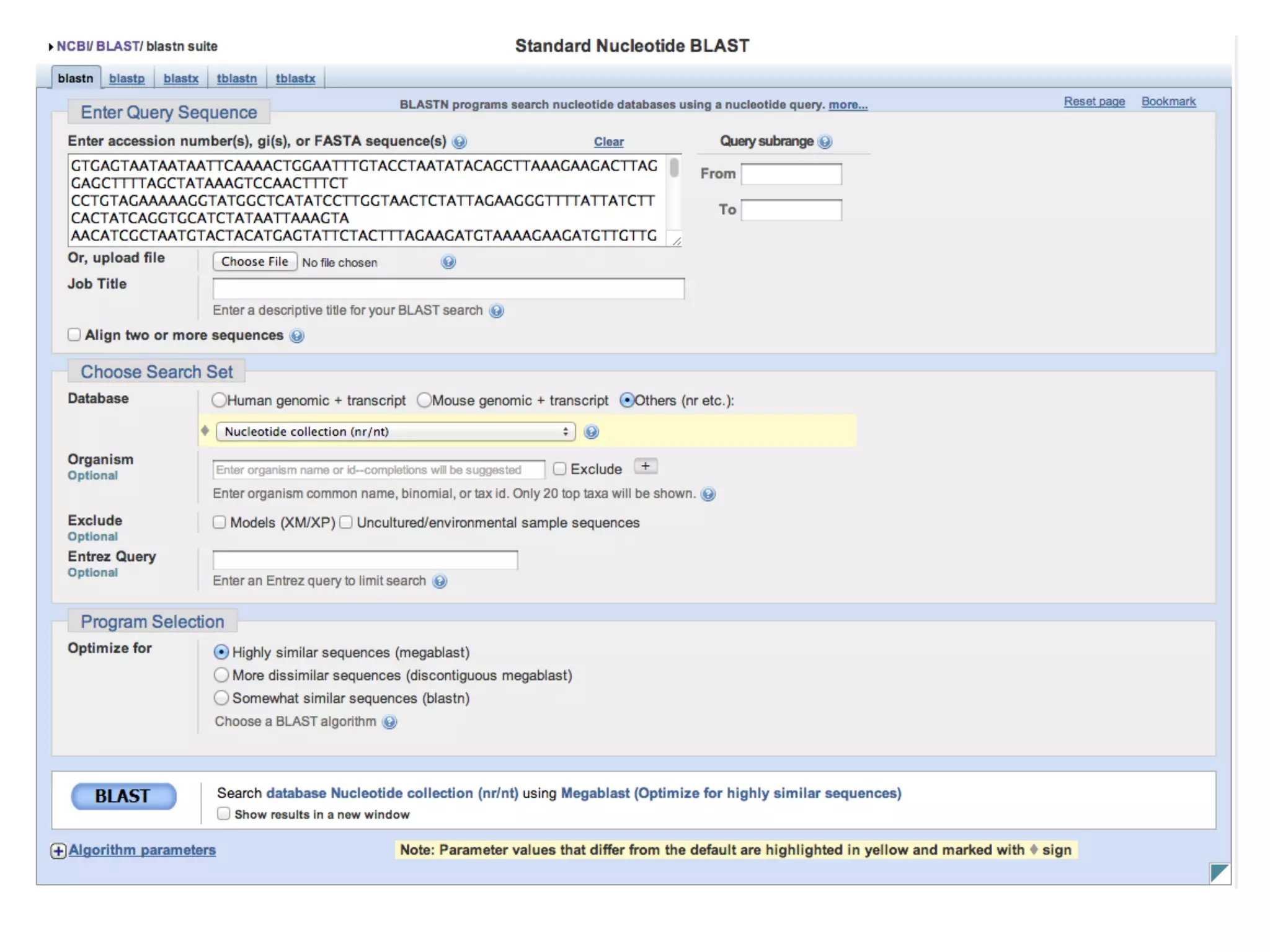
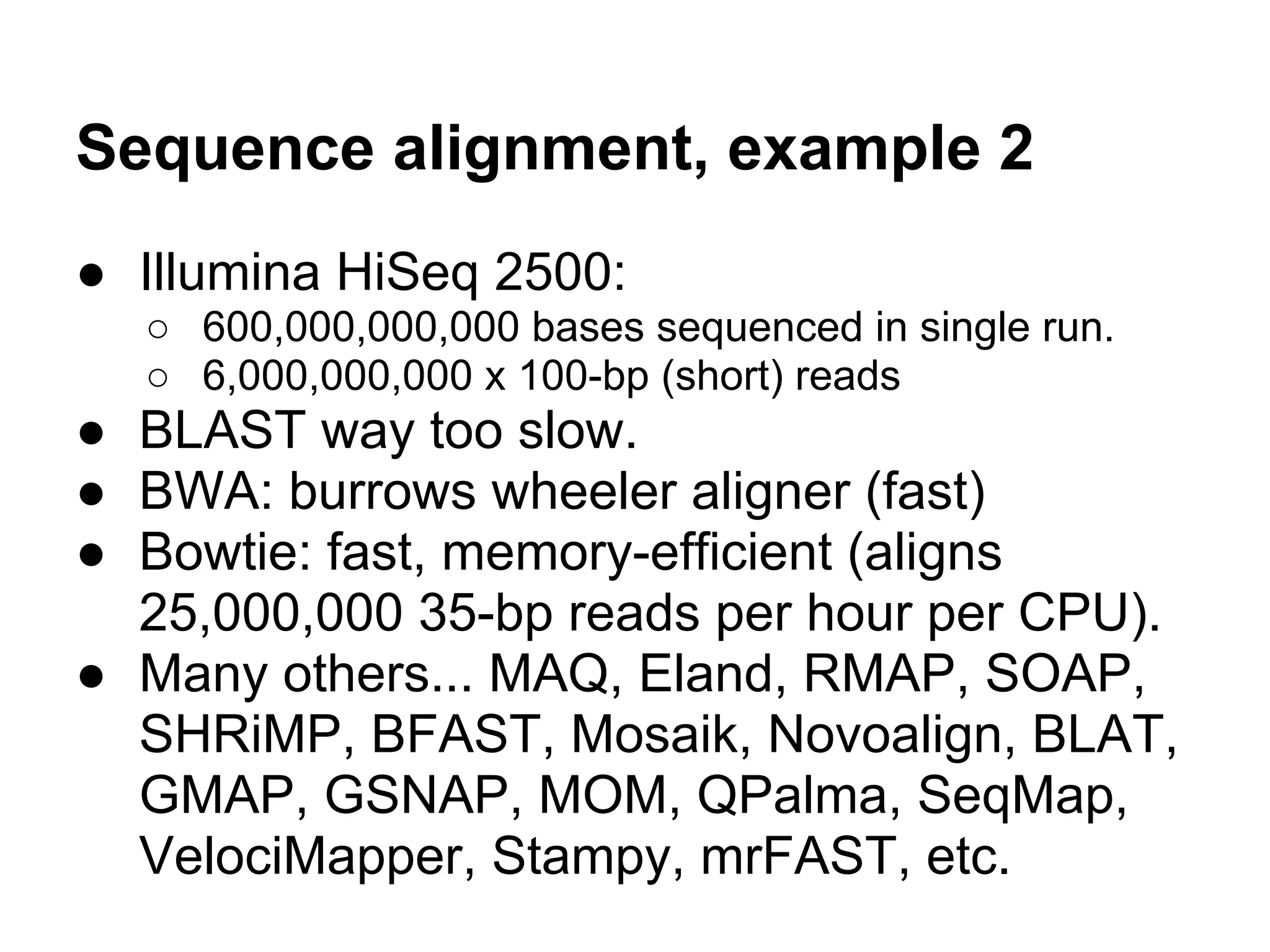
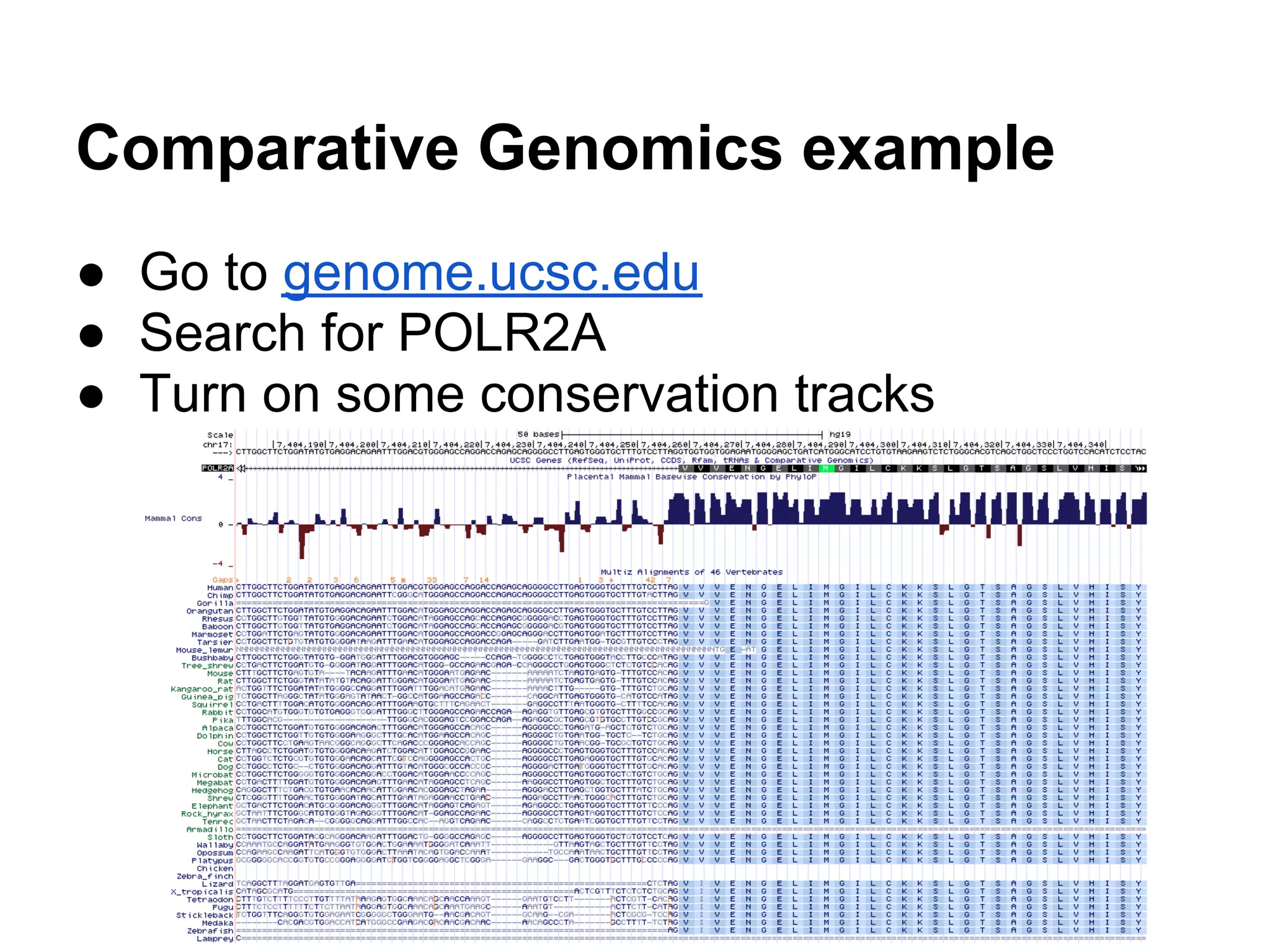
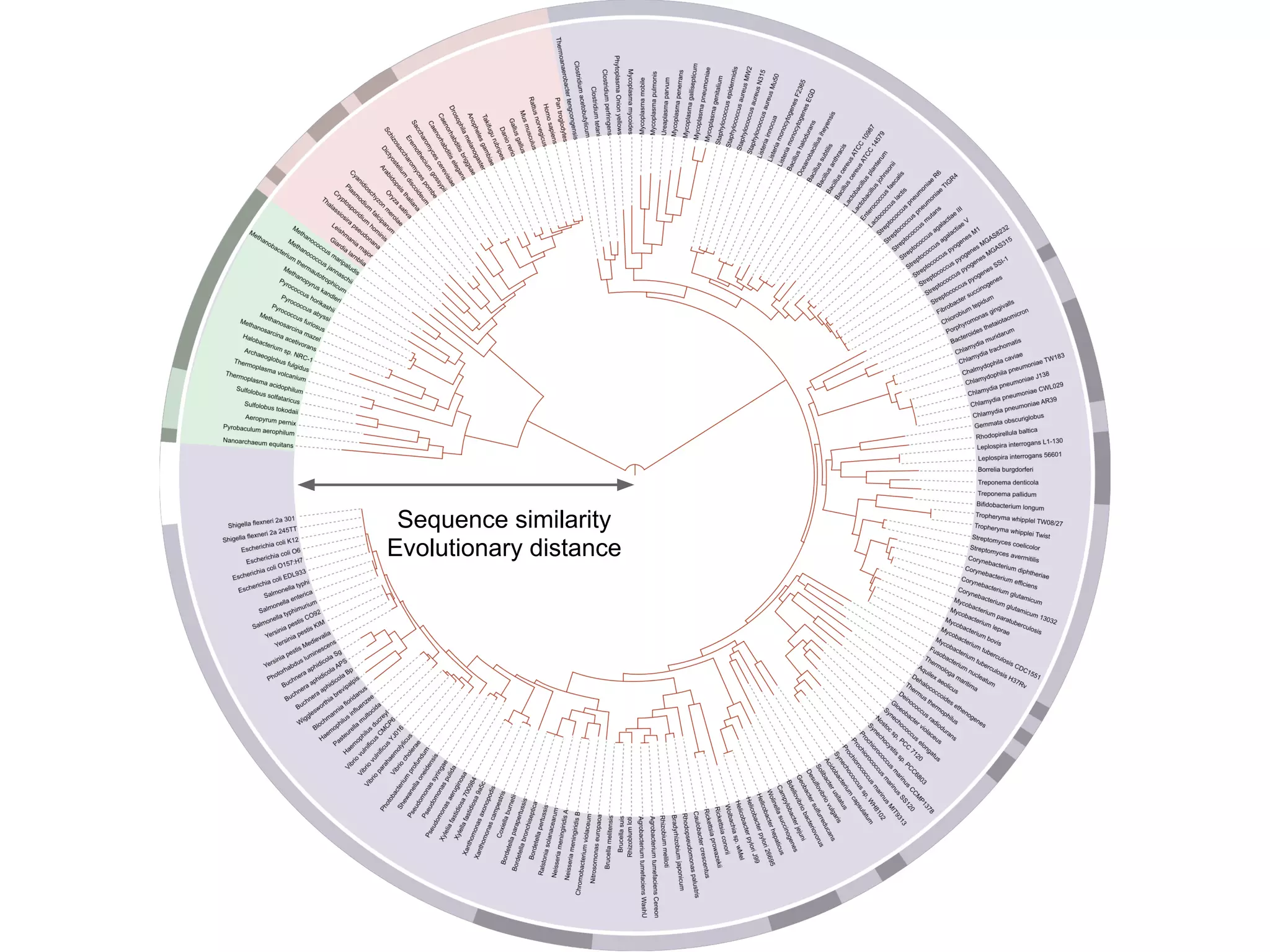
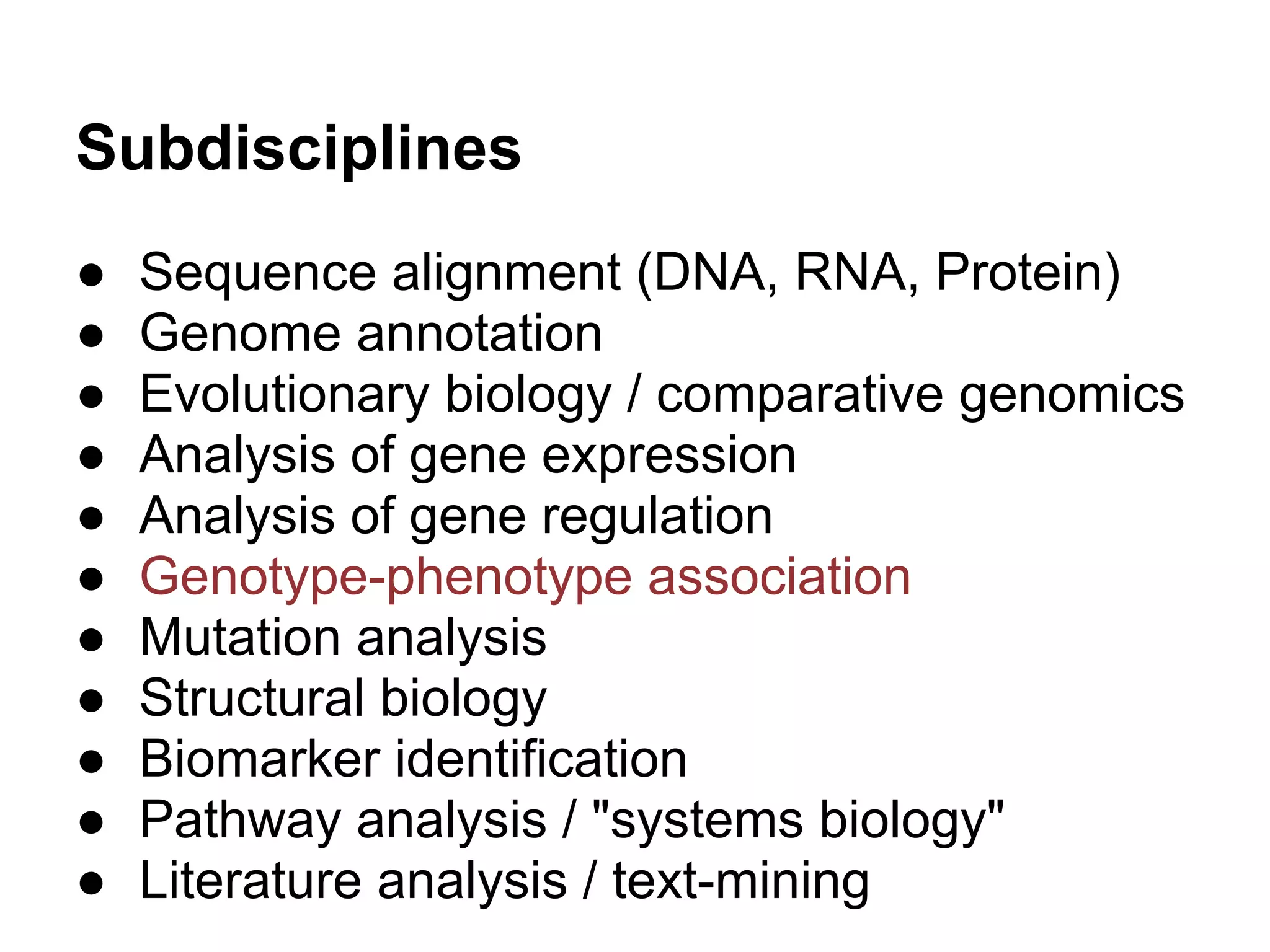
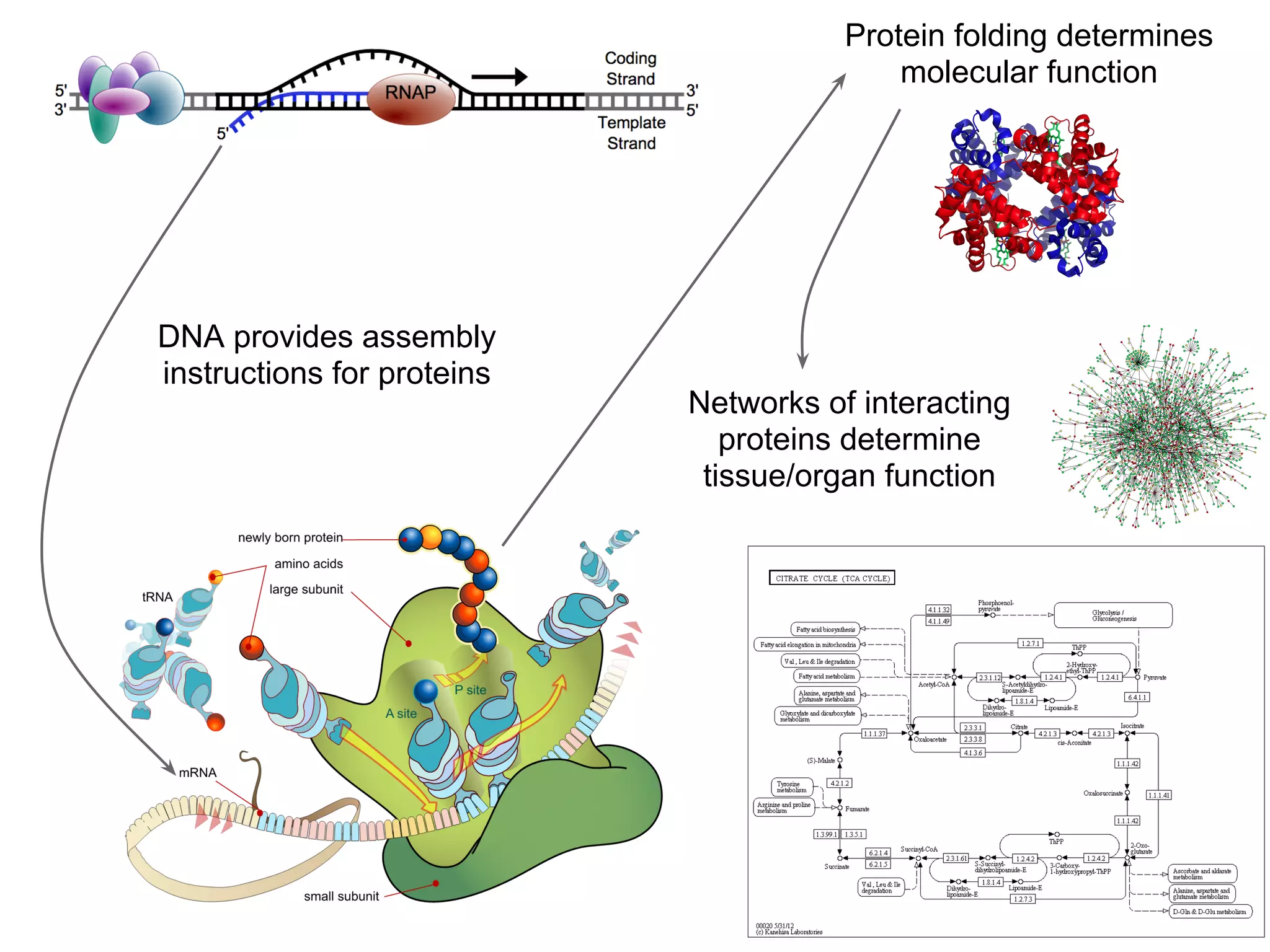
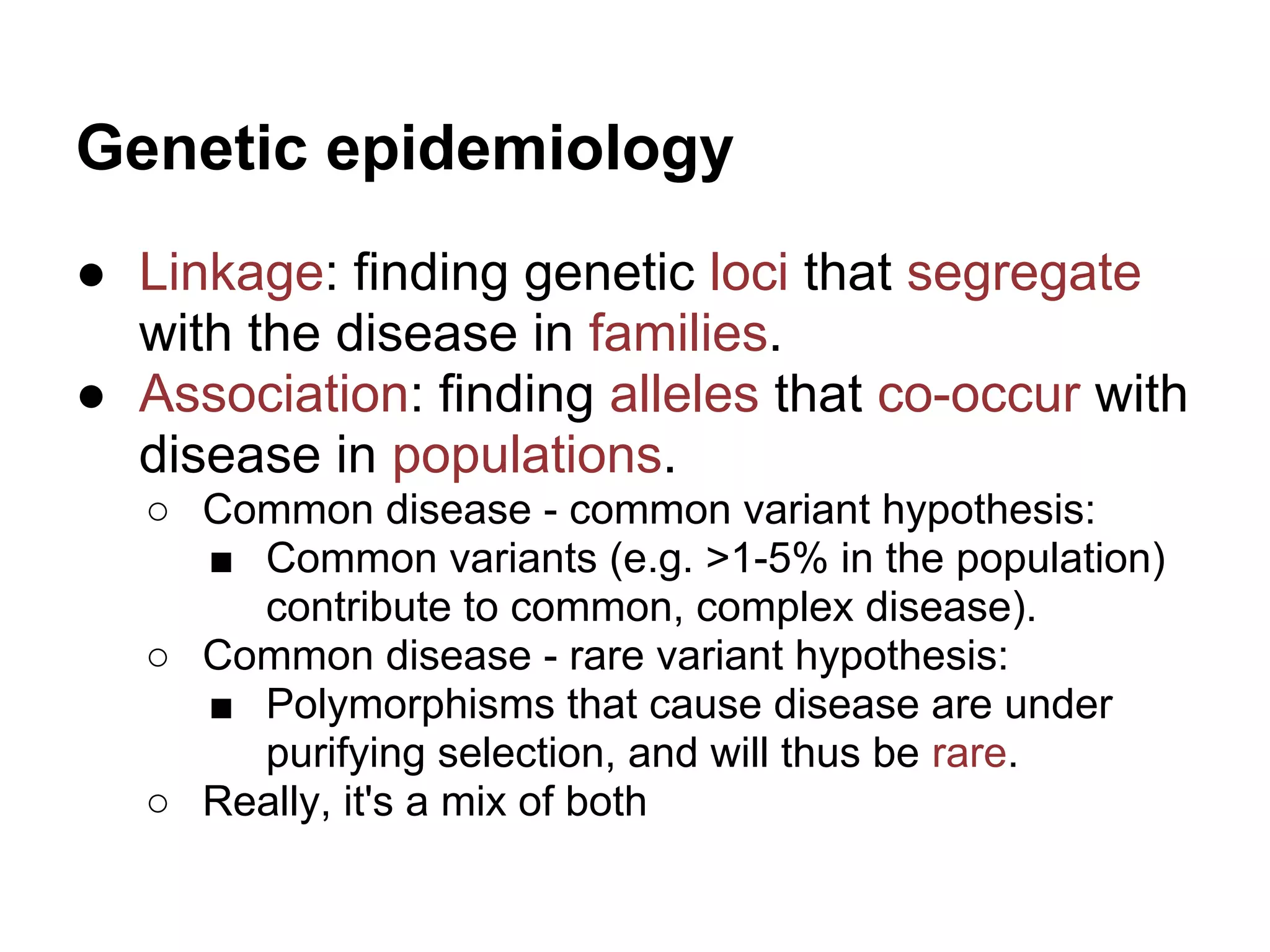
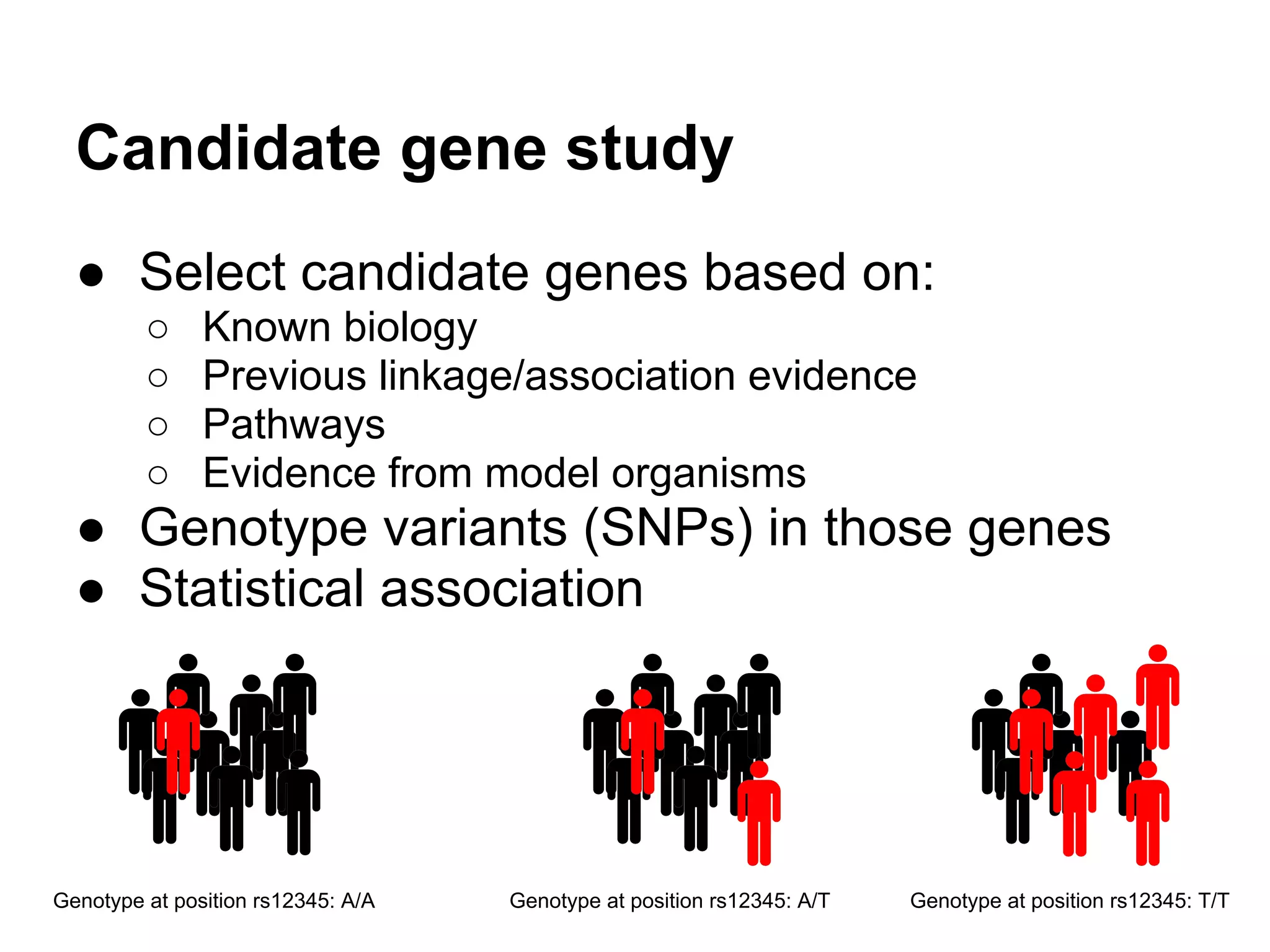
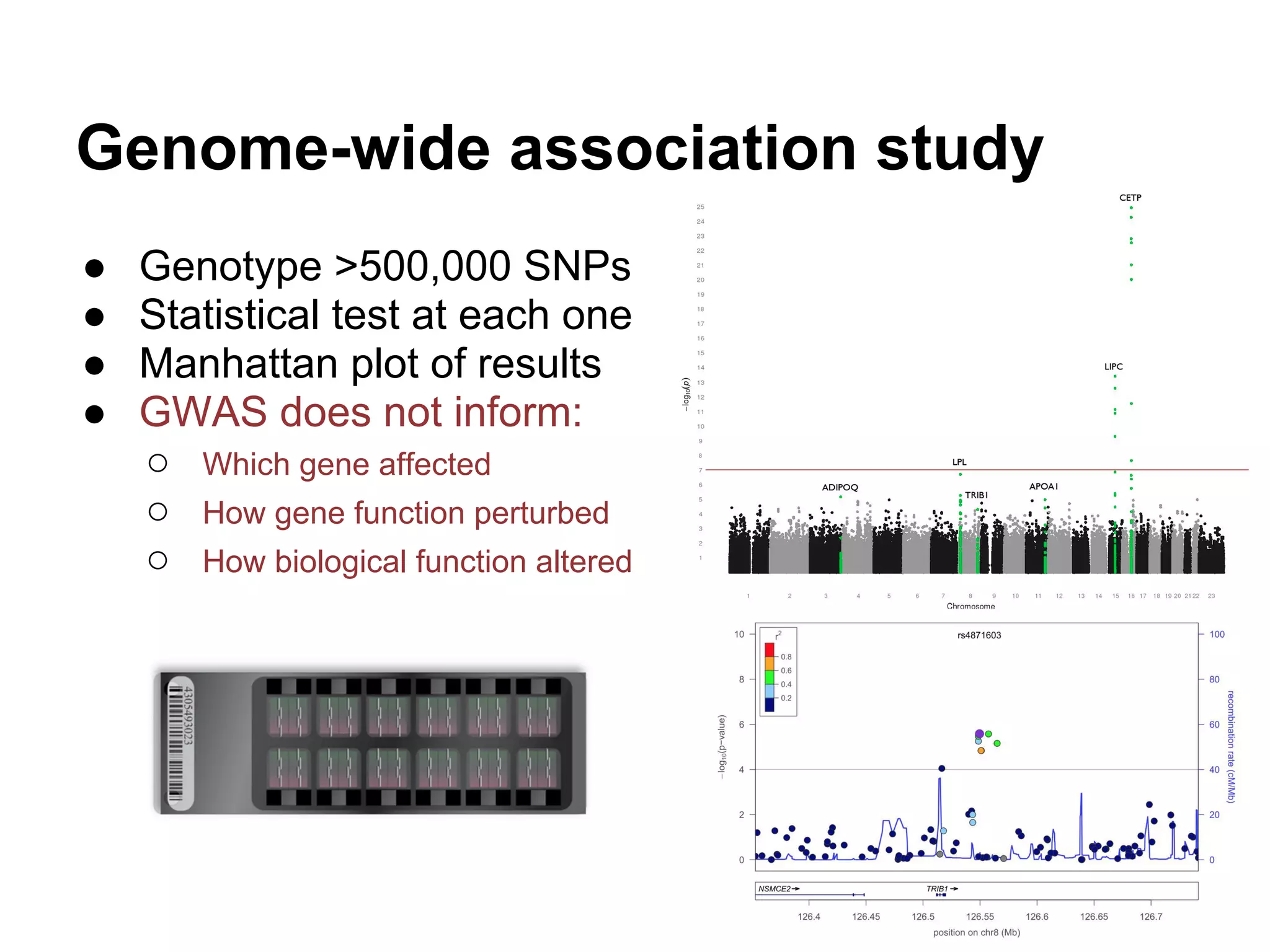
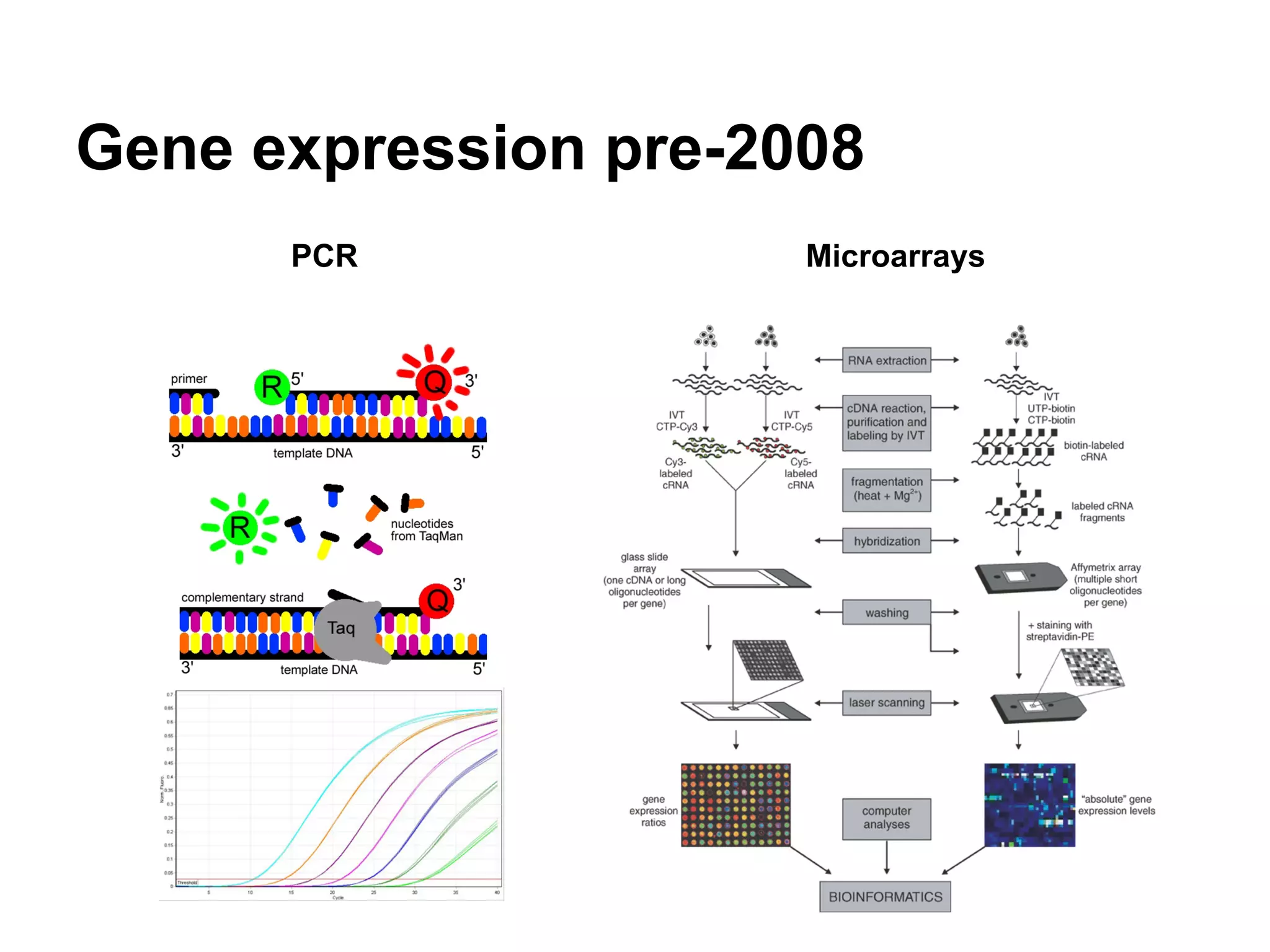
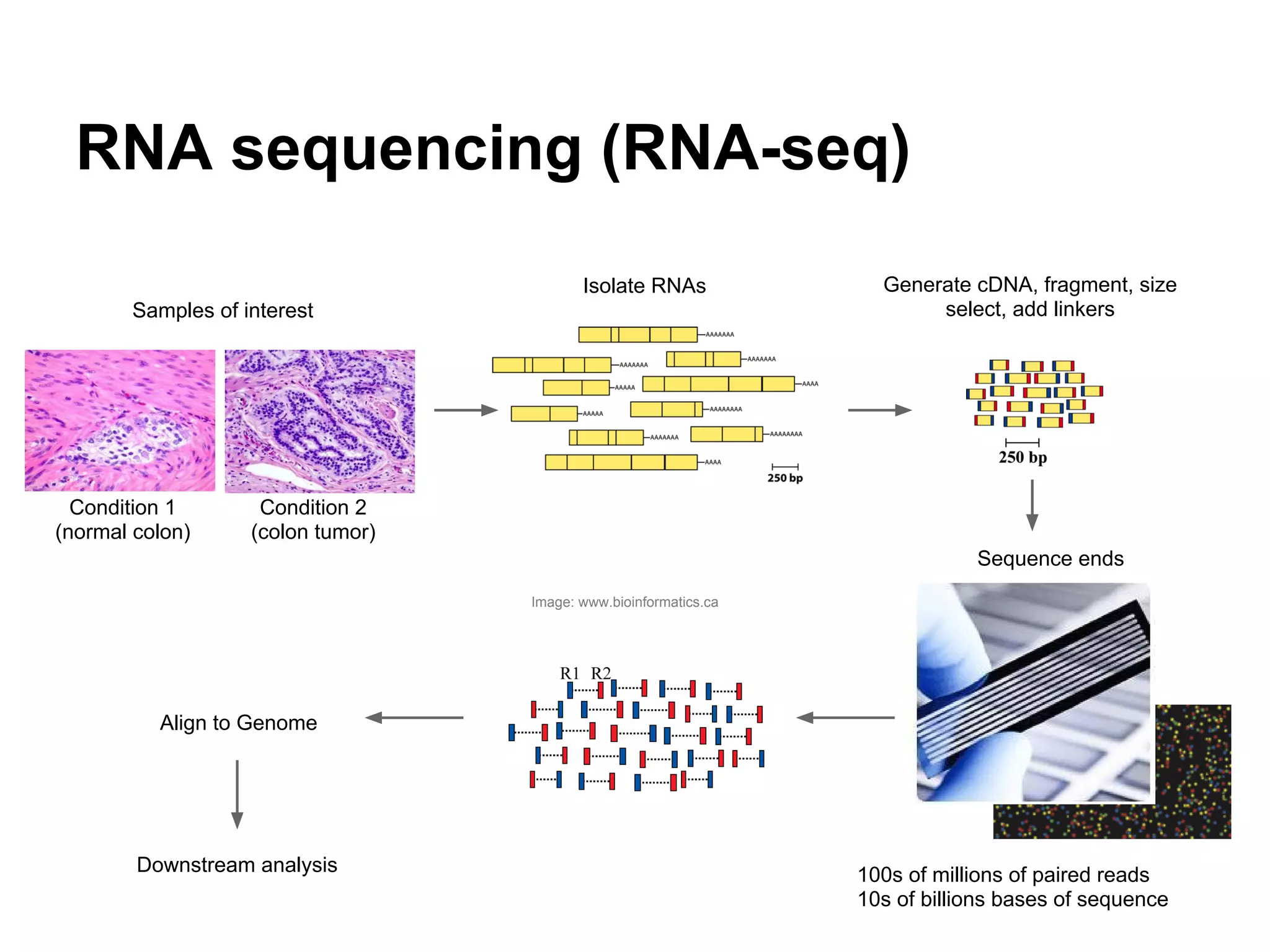
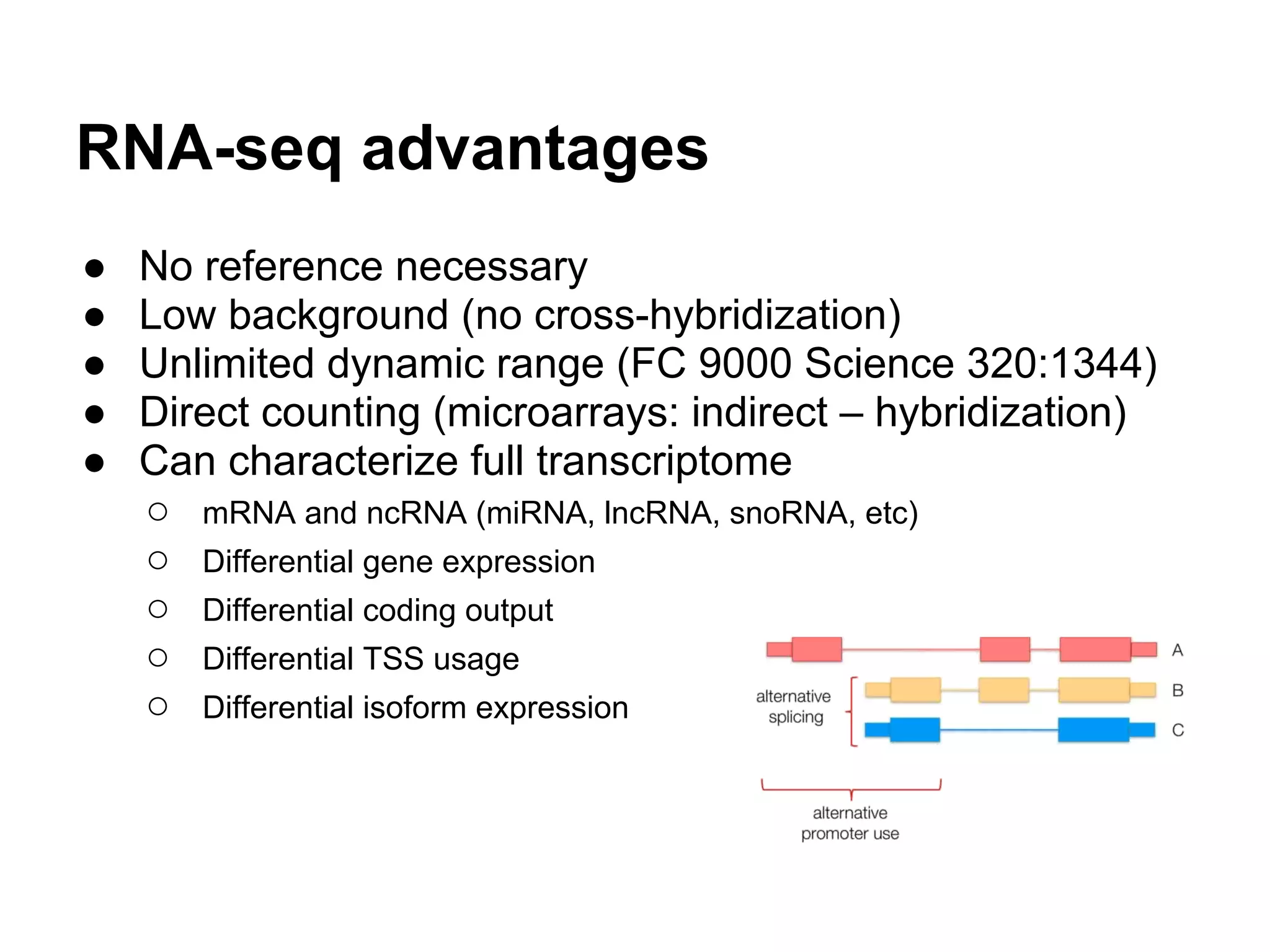
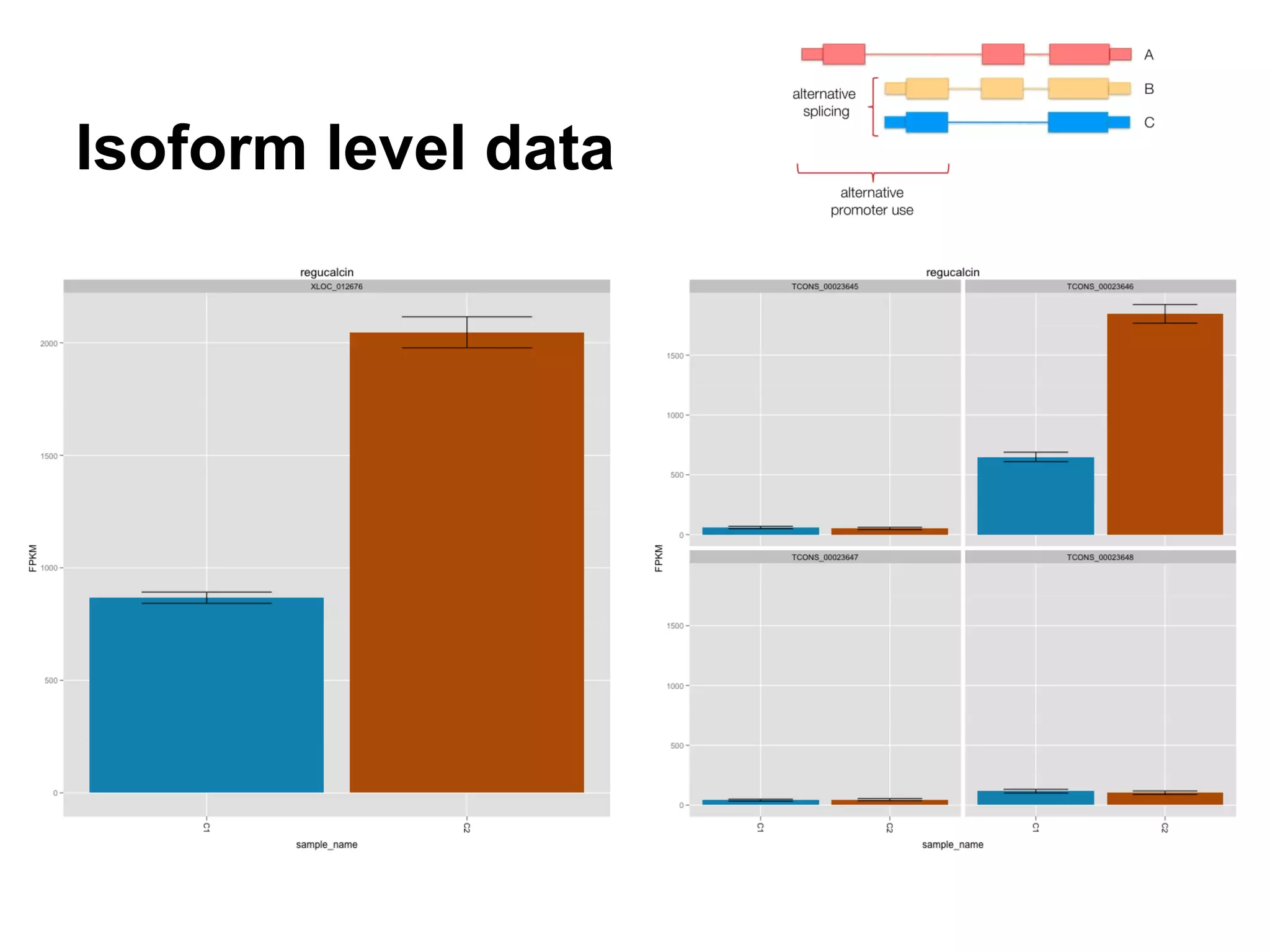
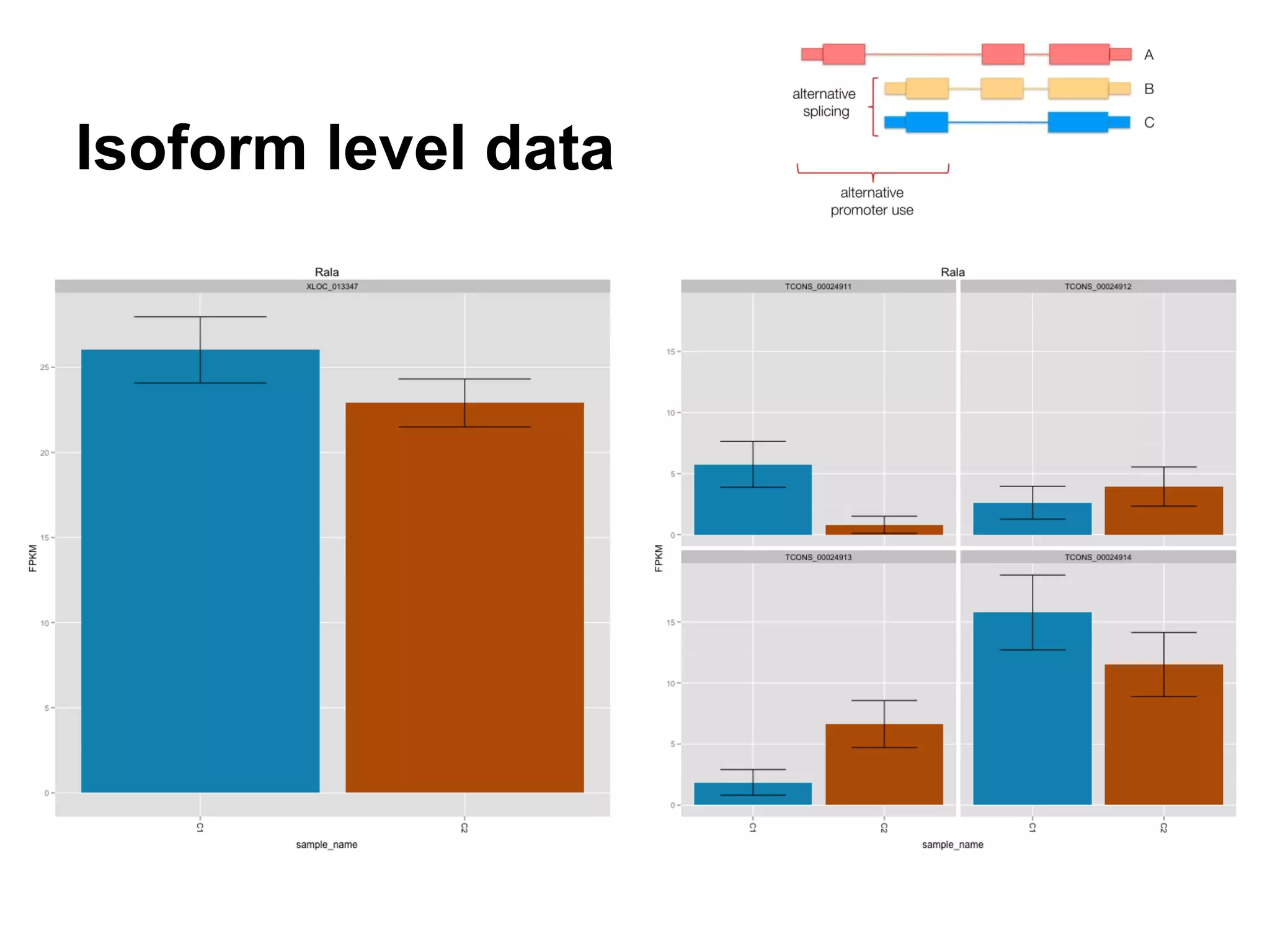
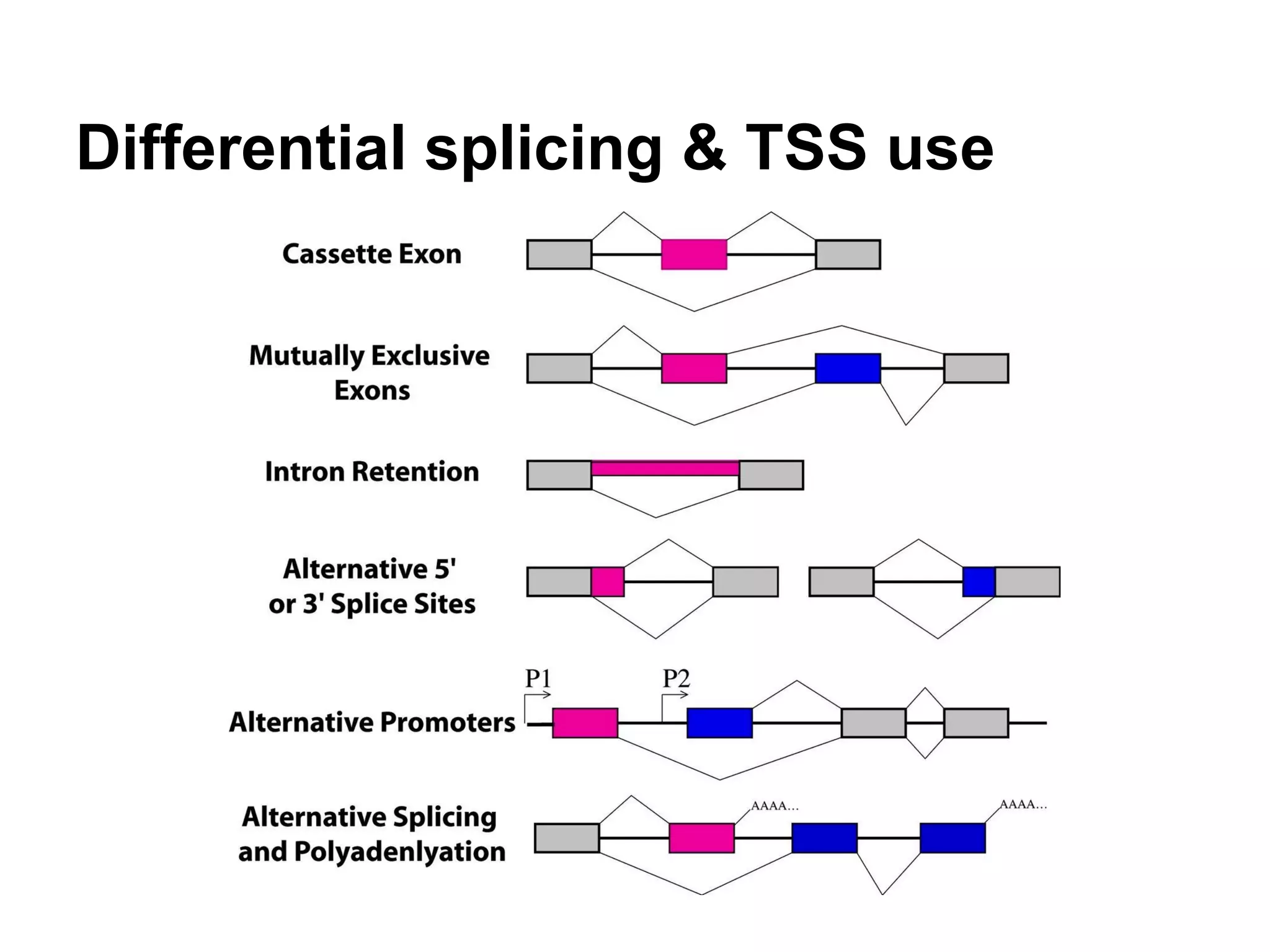
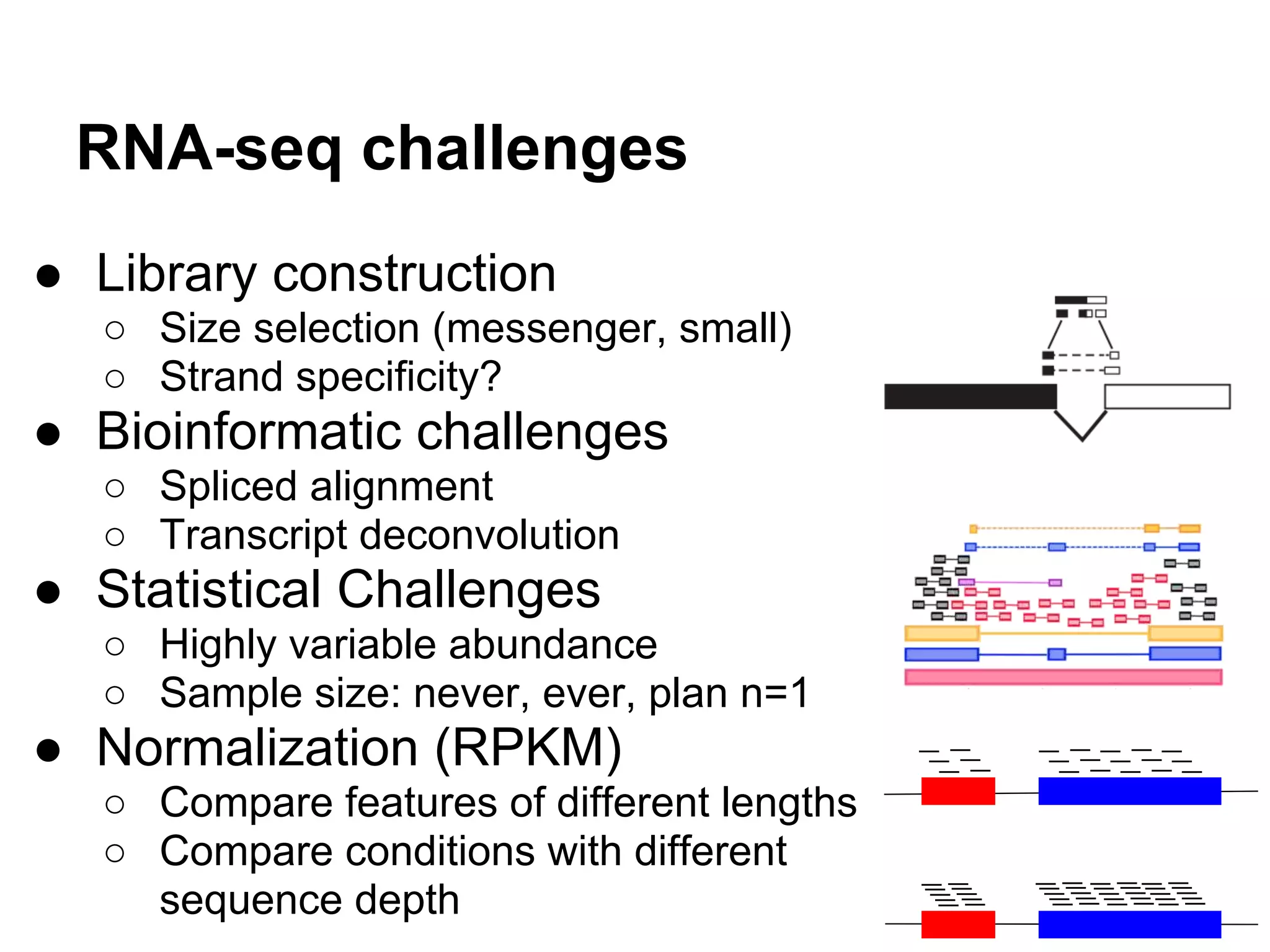
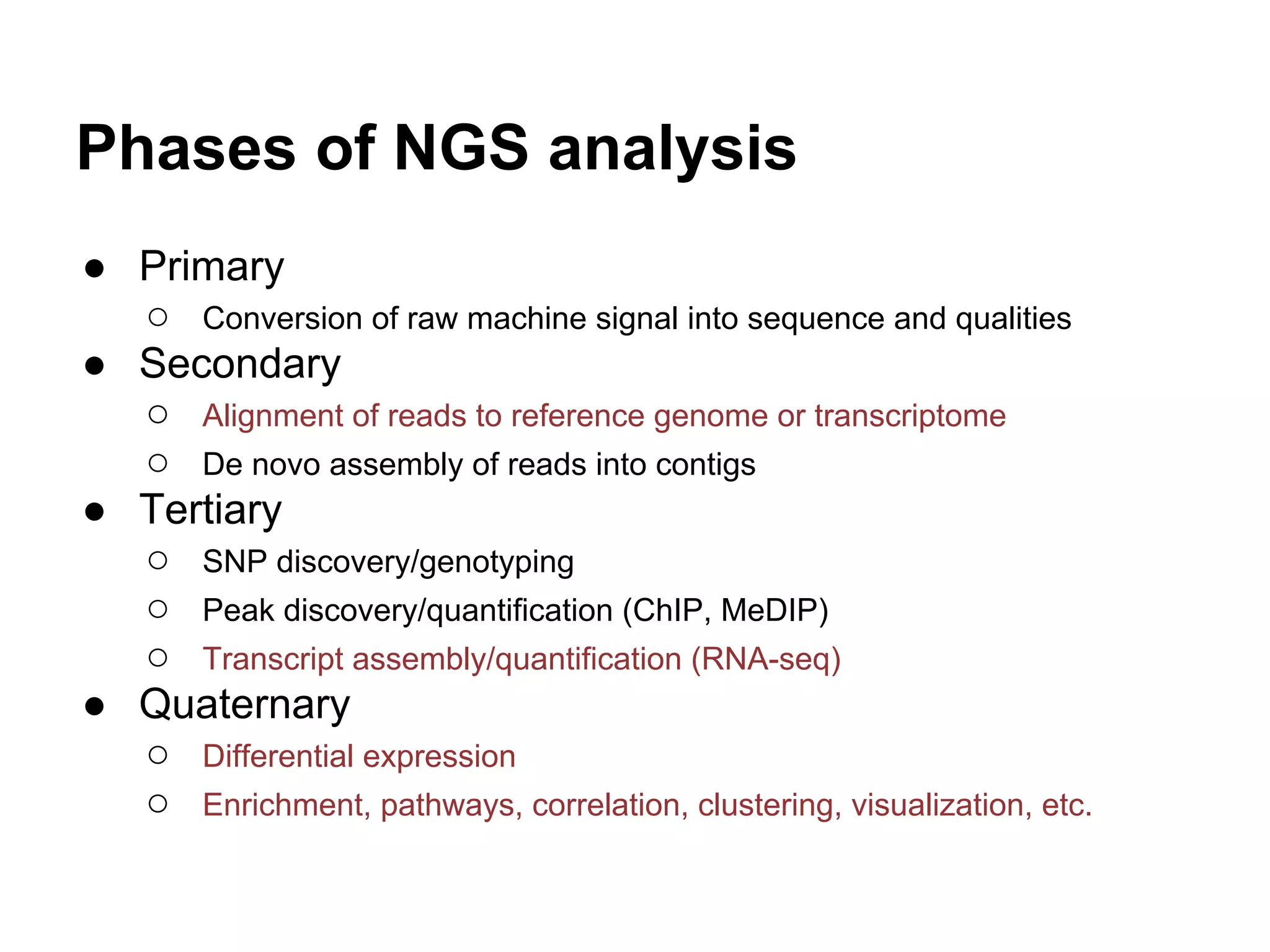
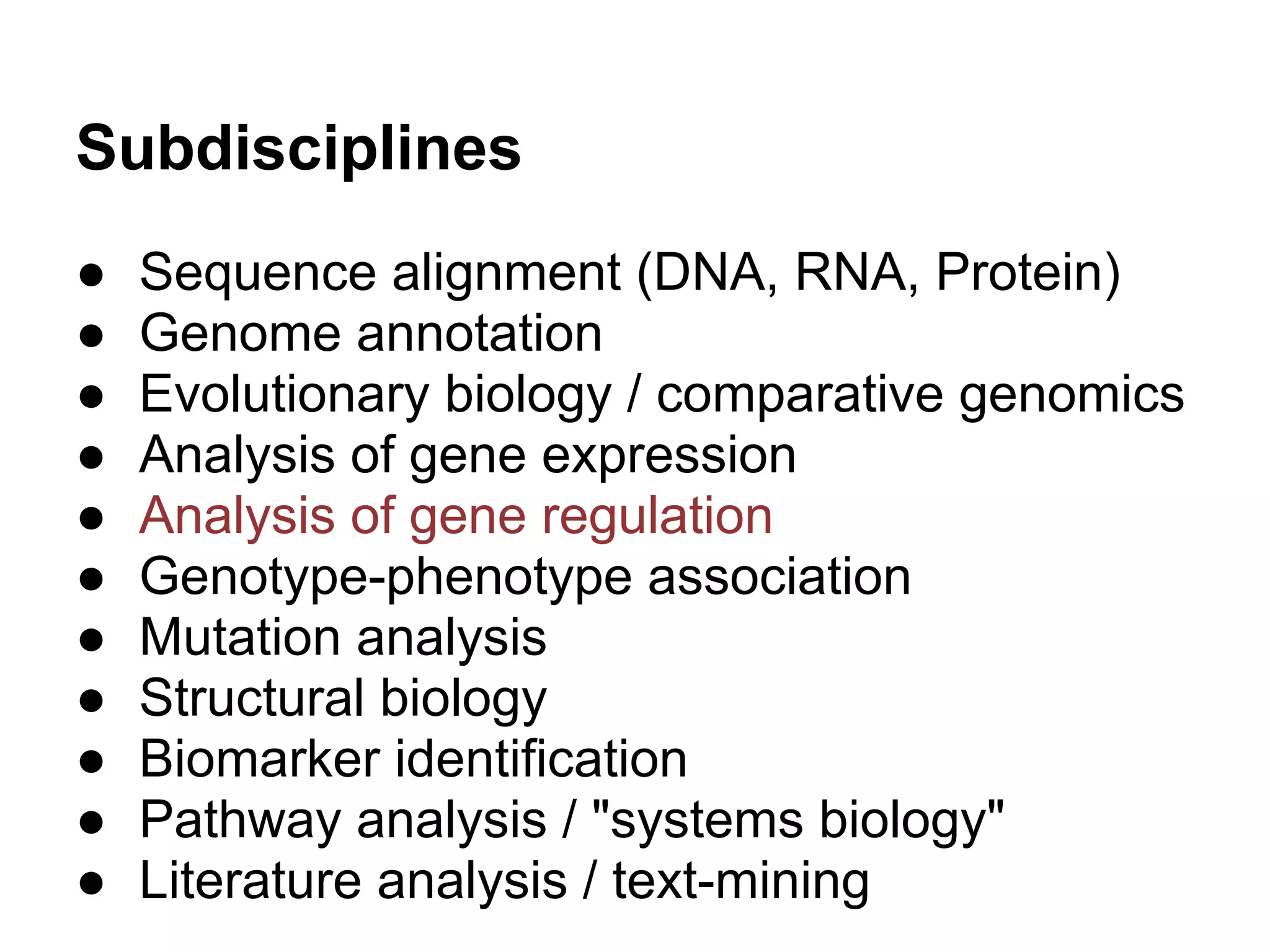
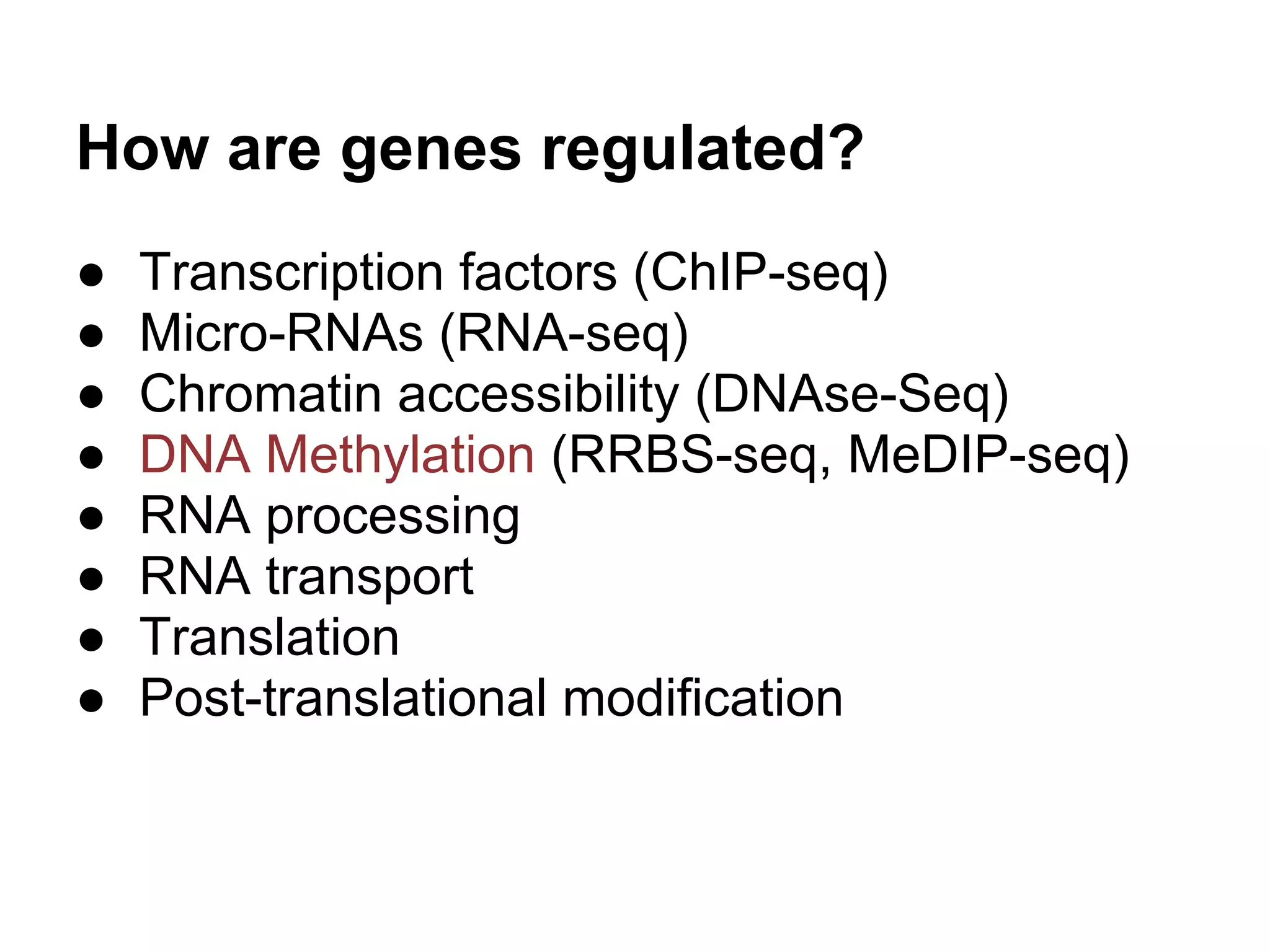
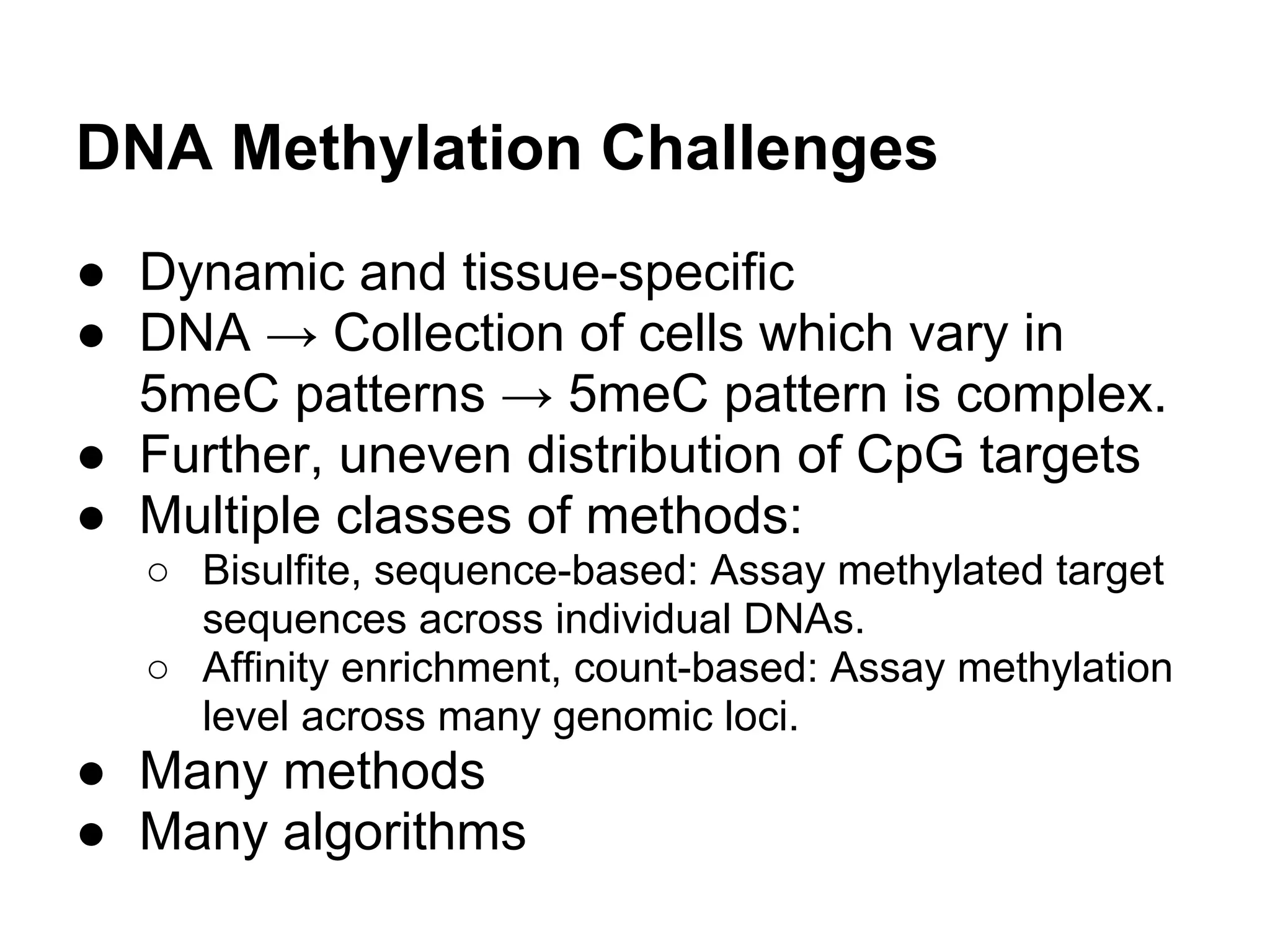
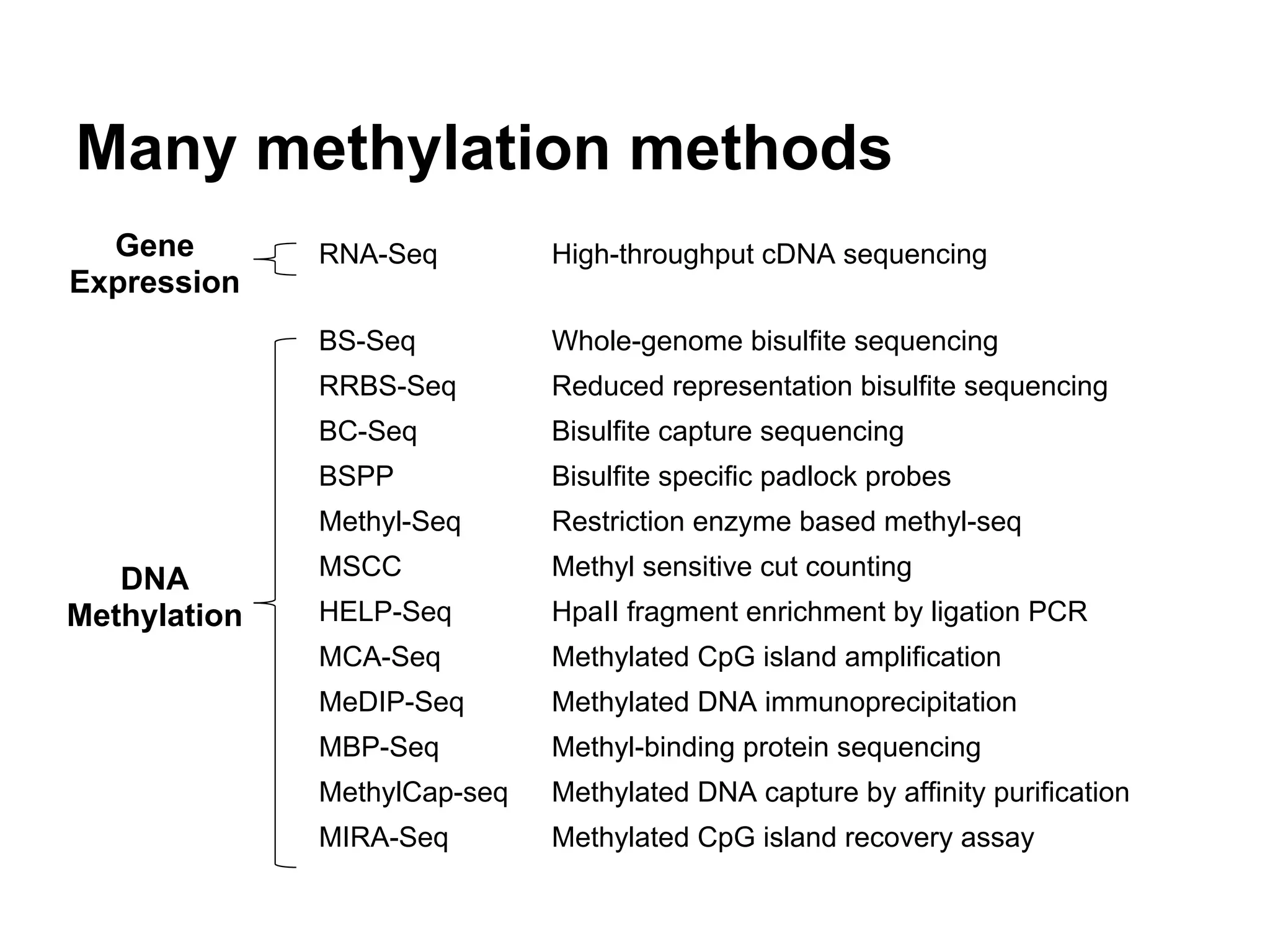
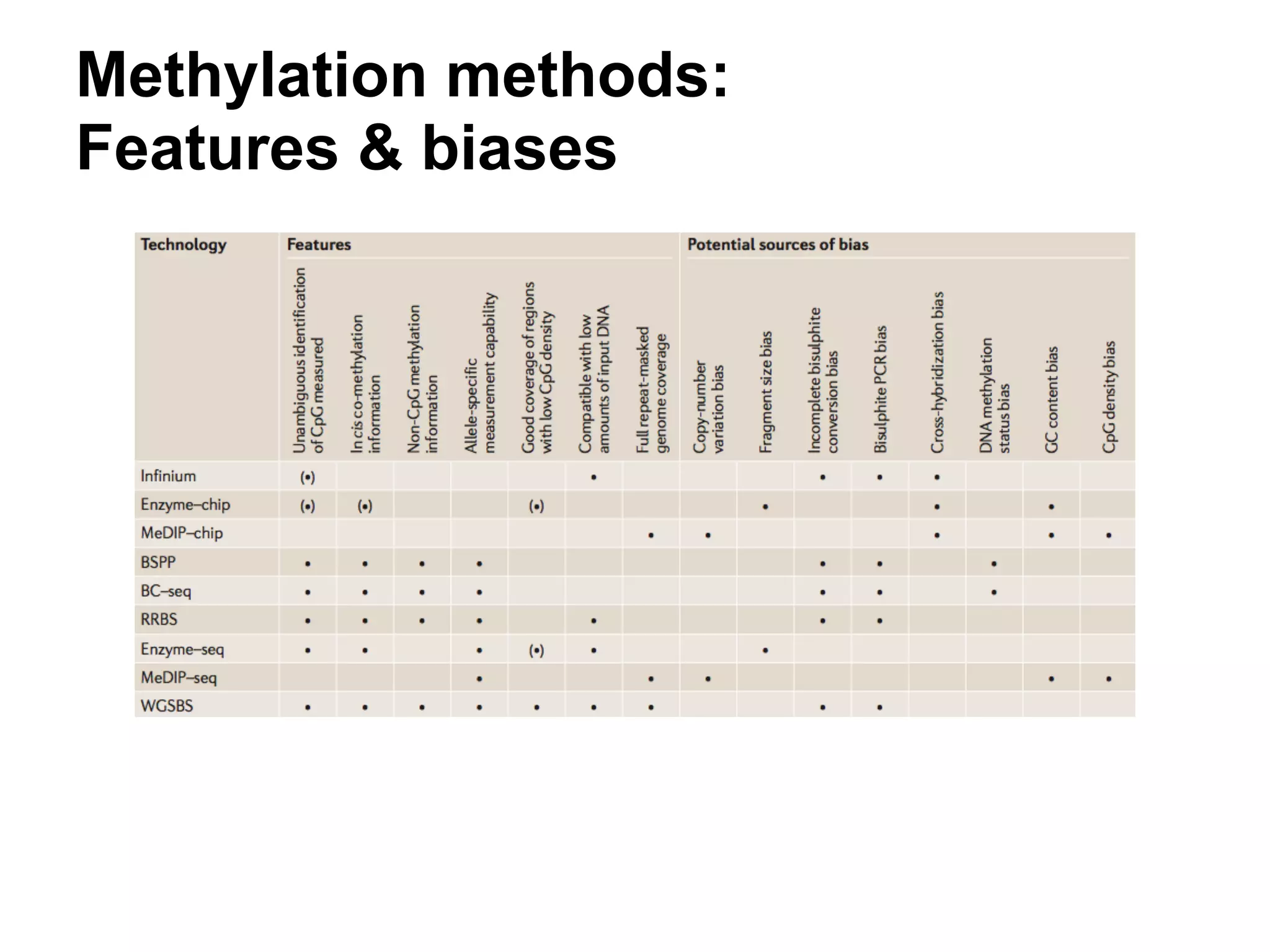
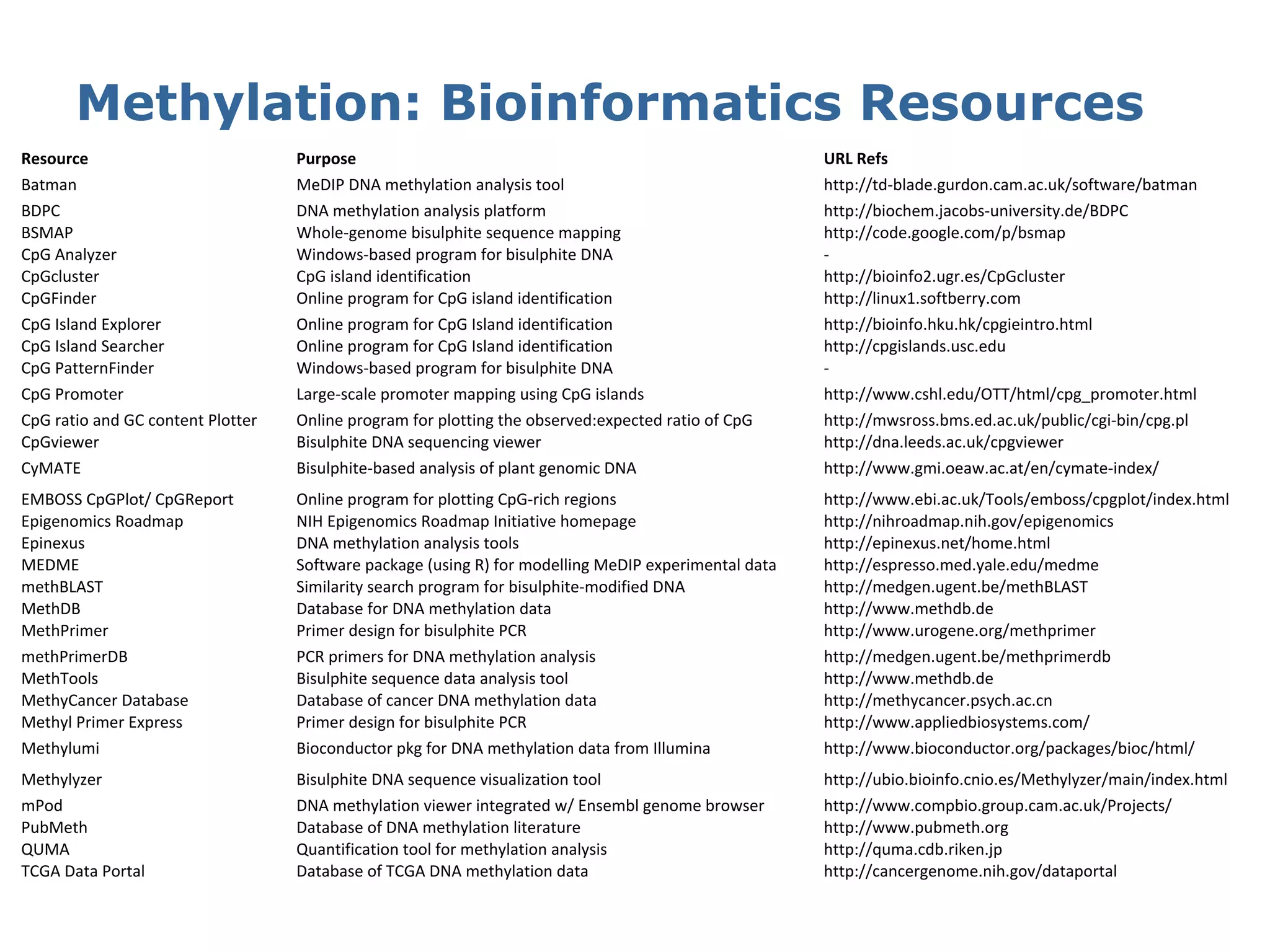
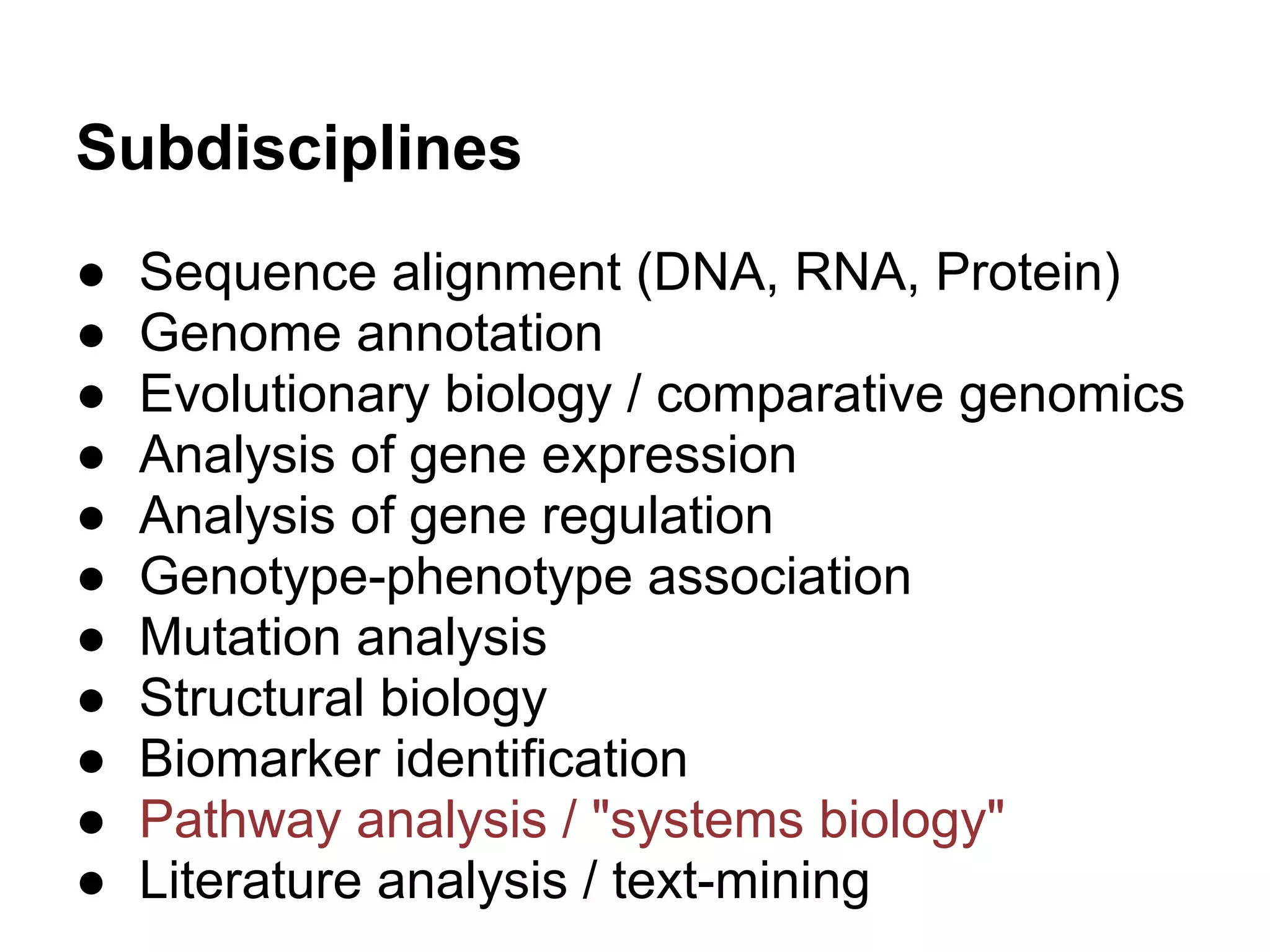
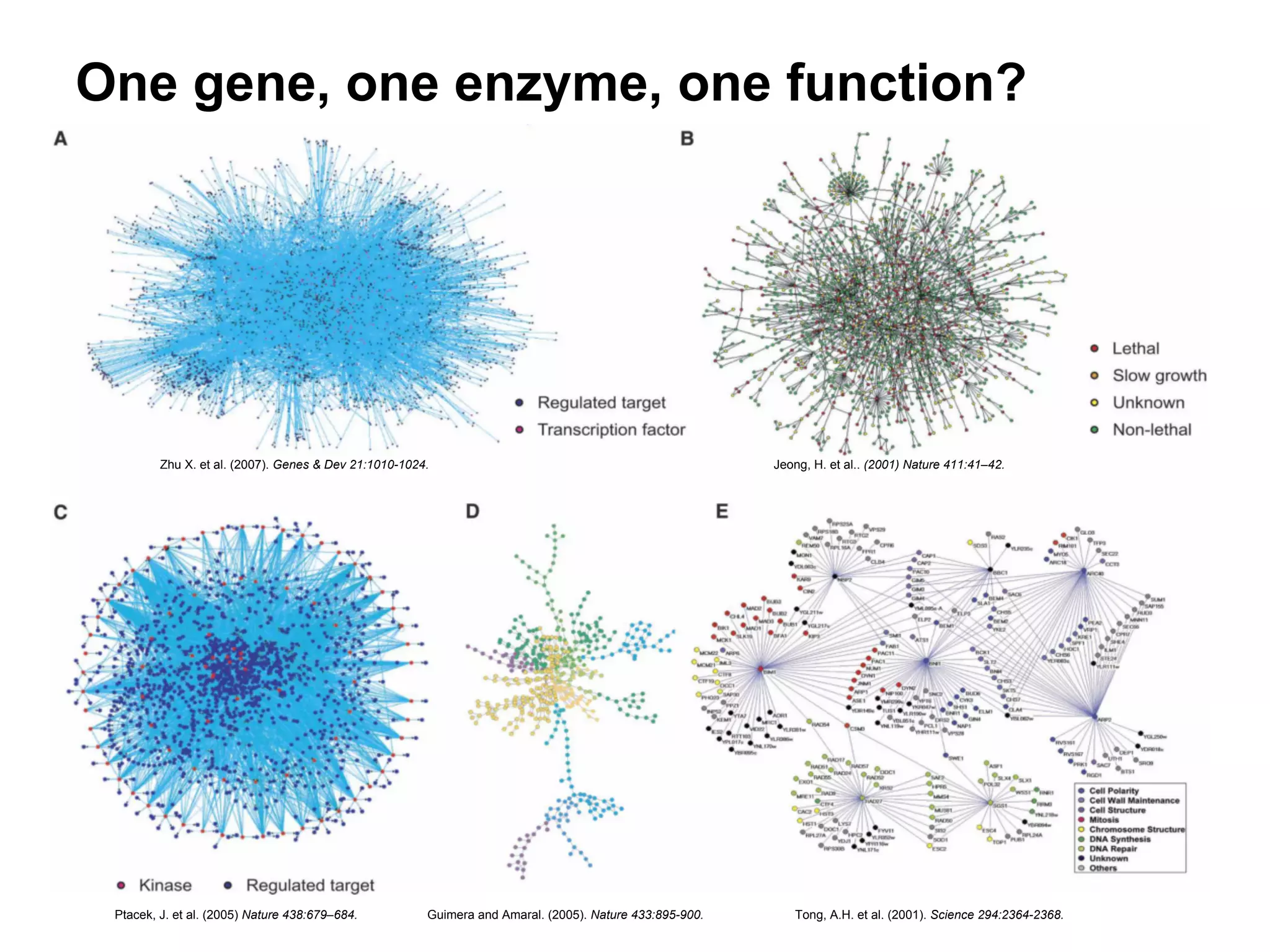
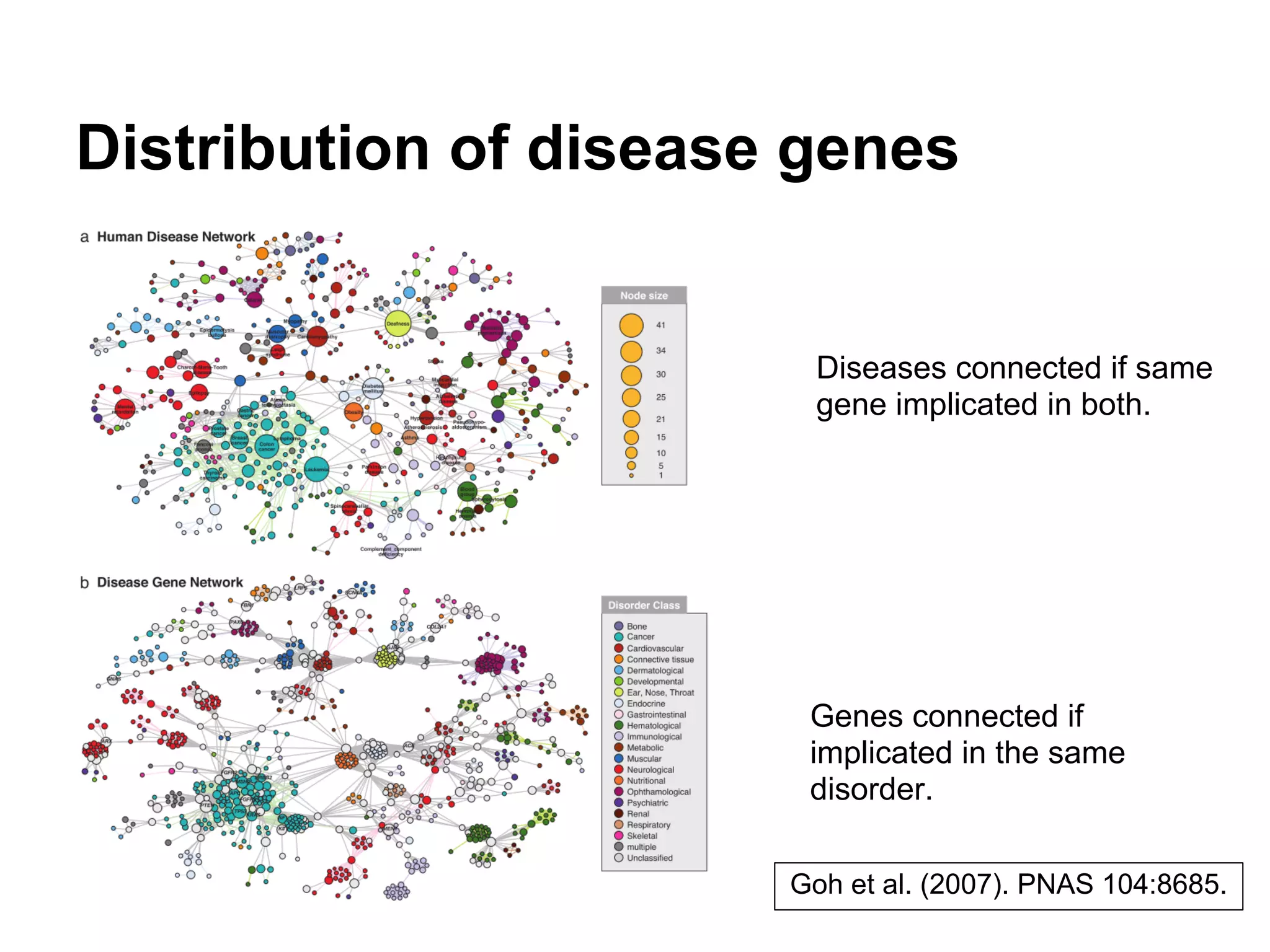
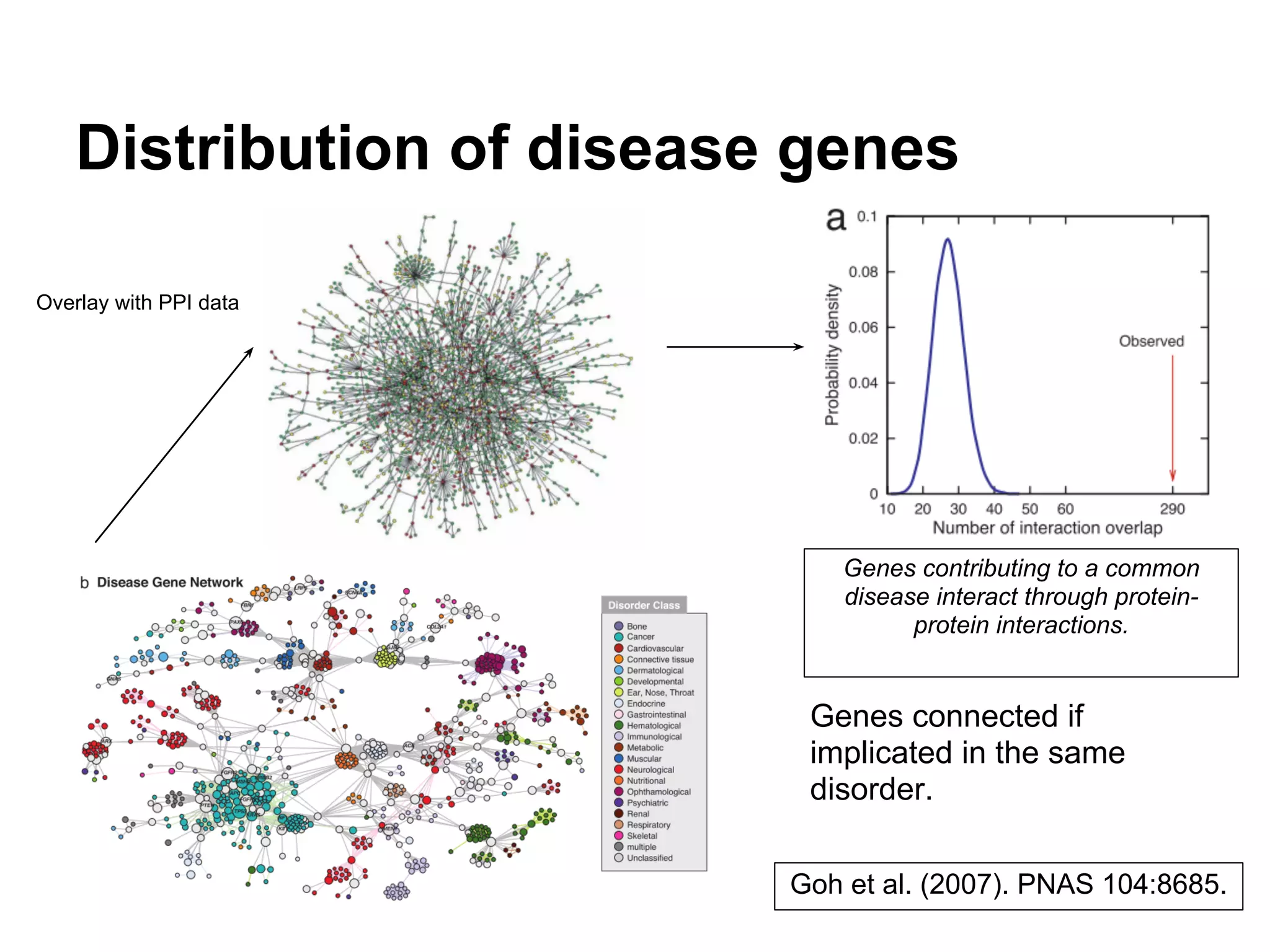
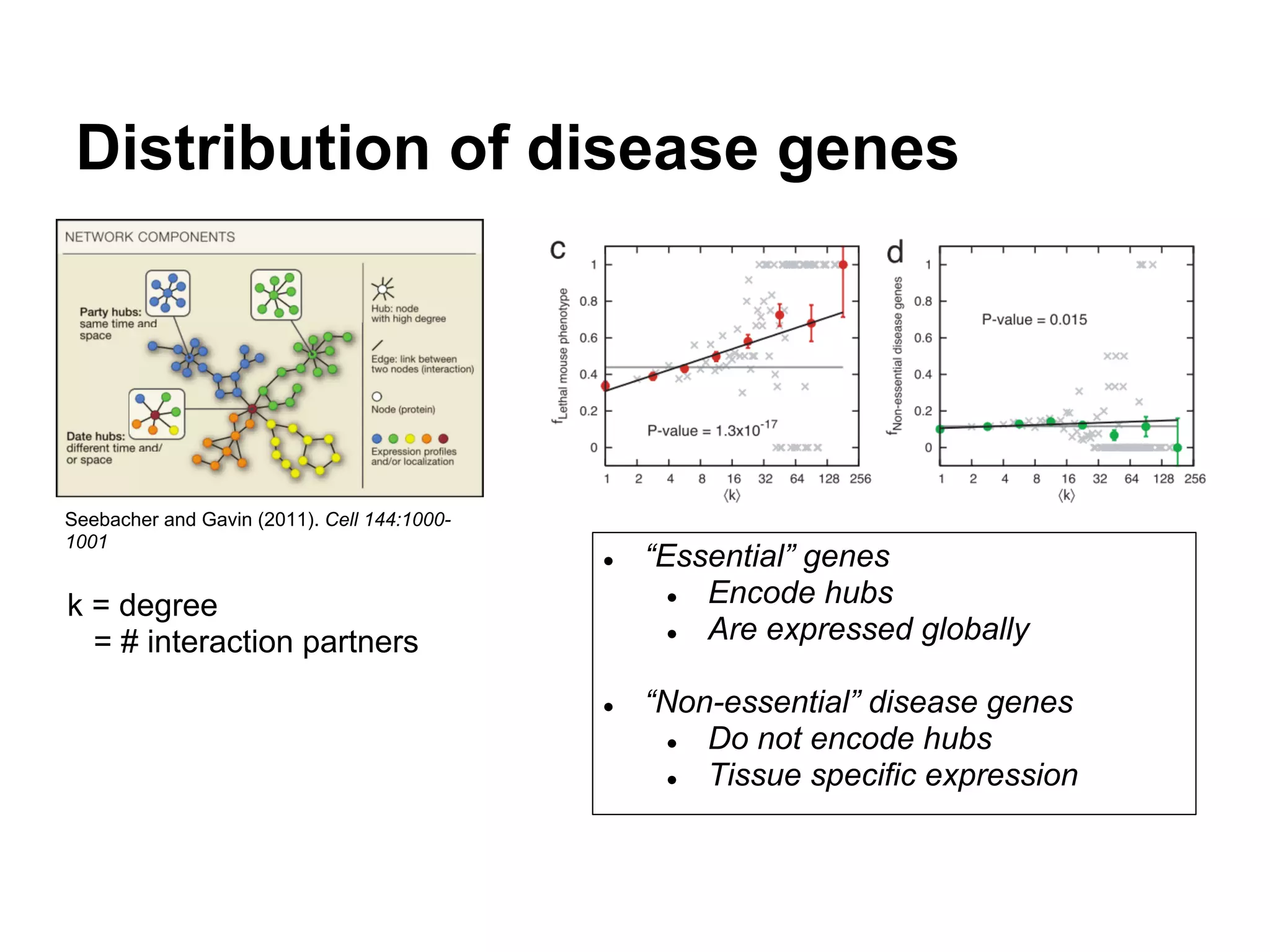
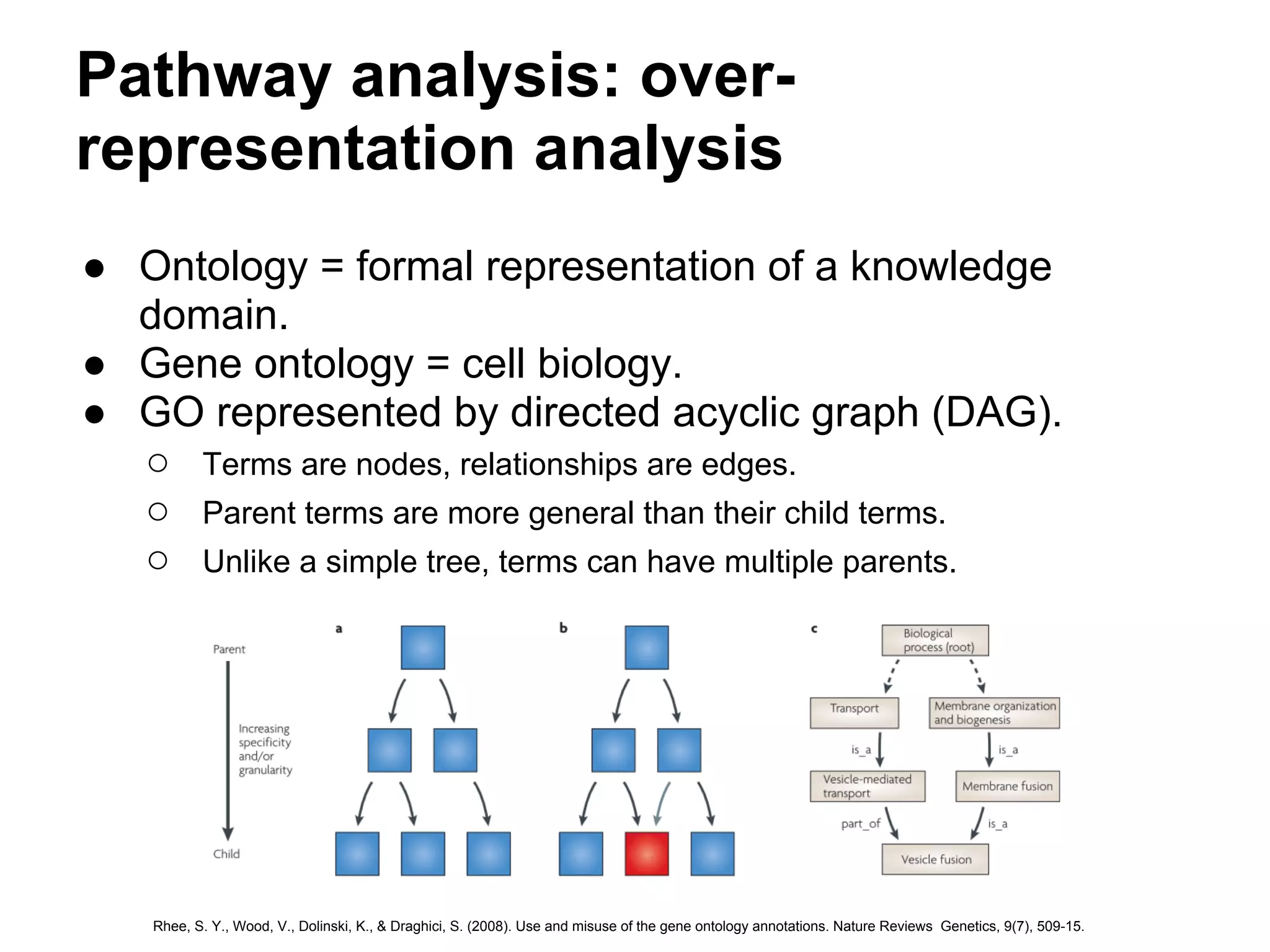
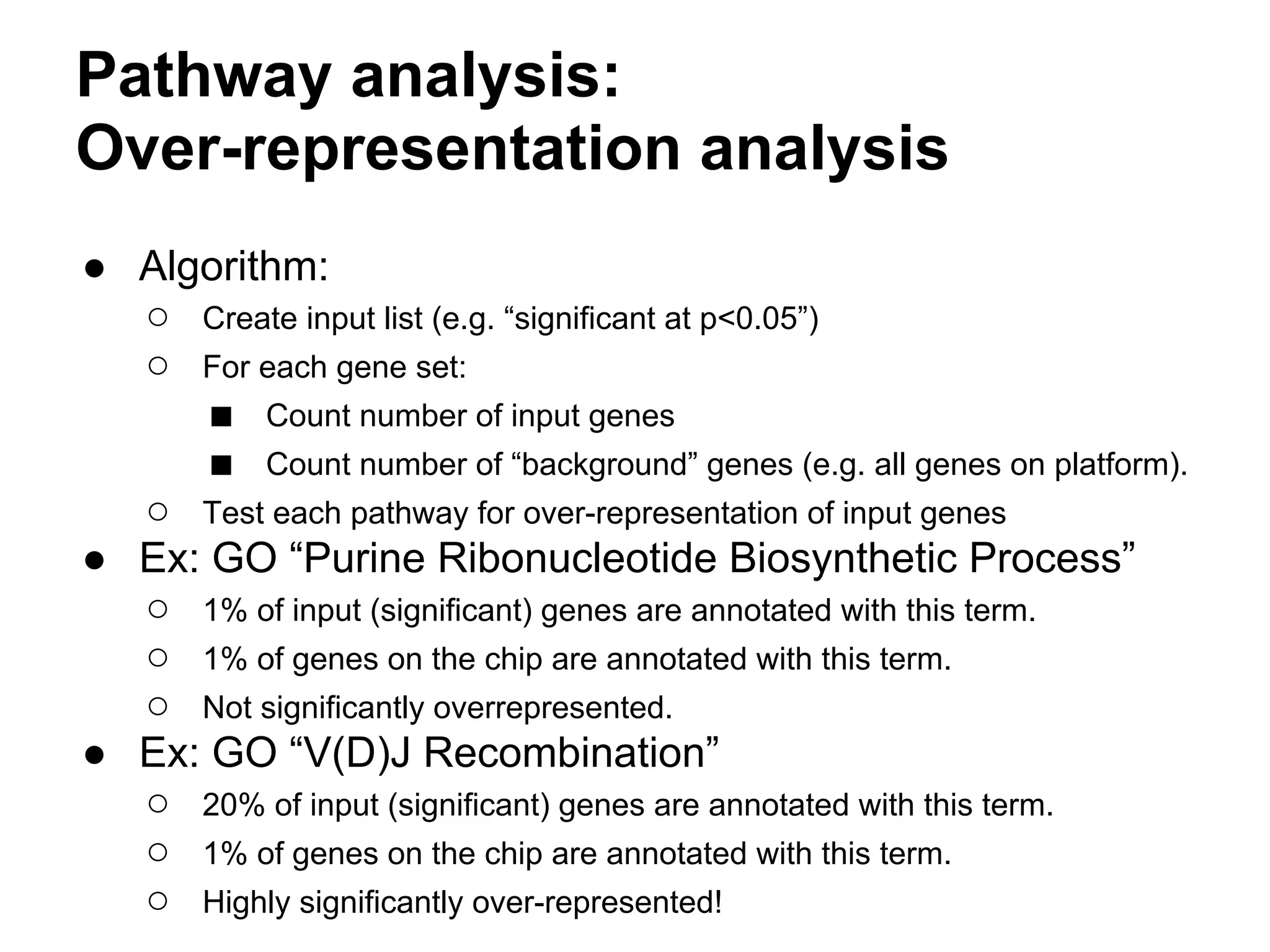
This document provides an introduction to the field of bioinformatics. It discusses how bioinformatics originated from the need to collect, annotate, and analyze biological sequence data. One pioneering researcher, Margaret Dayhoff, collected all known protein structures and sequences in 1965 and developed early algorithms to compare sequences and study evolutionary relationships. The document outlines several subdisciplines of bioinformatics including sequence alignment, genome annotation, analysis of gene expression, pathway analysis, and literature analysis. It provides examples of using tools like BLAST for sequence alignment and the UCSC Genome Browser for comparative genomics analysis.