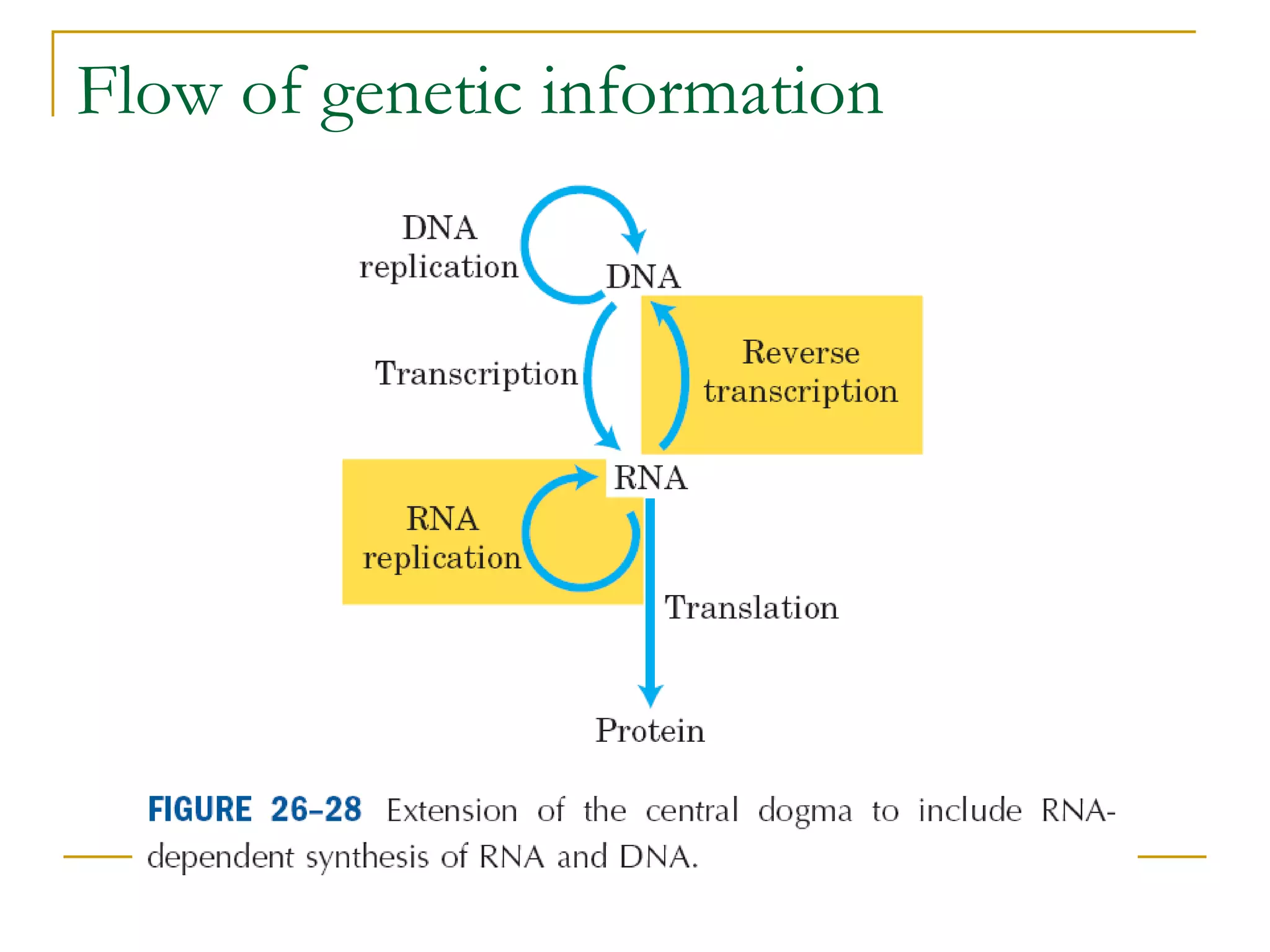
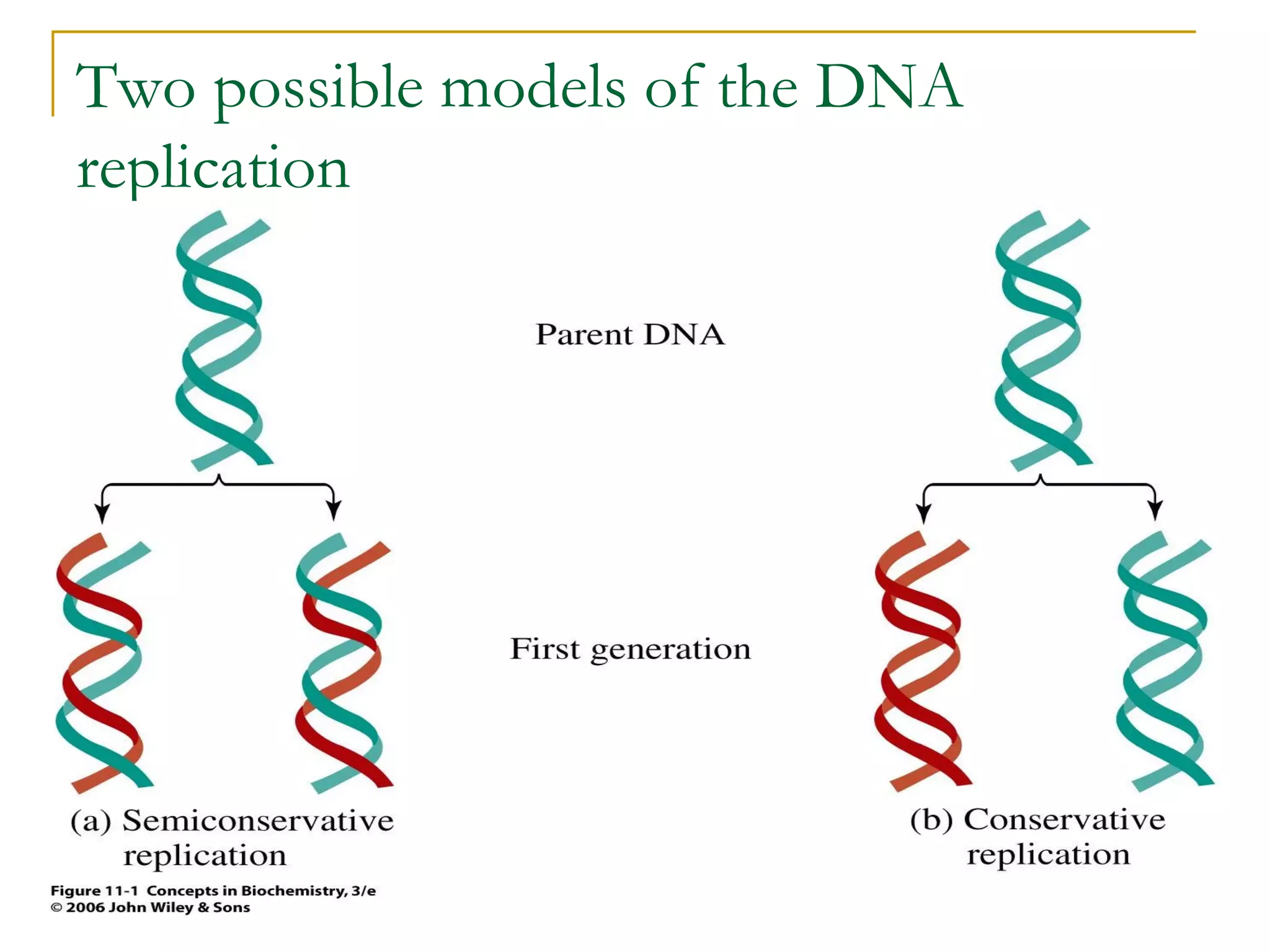
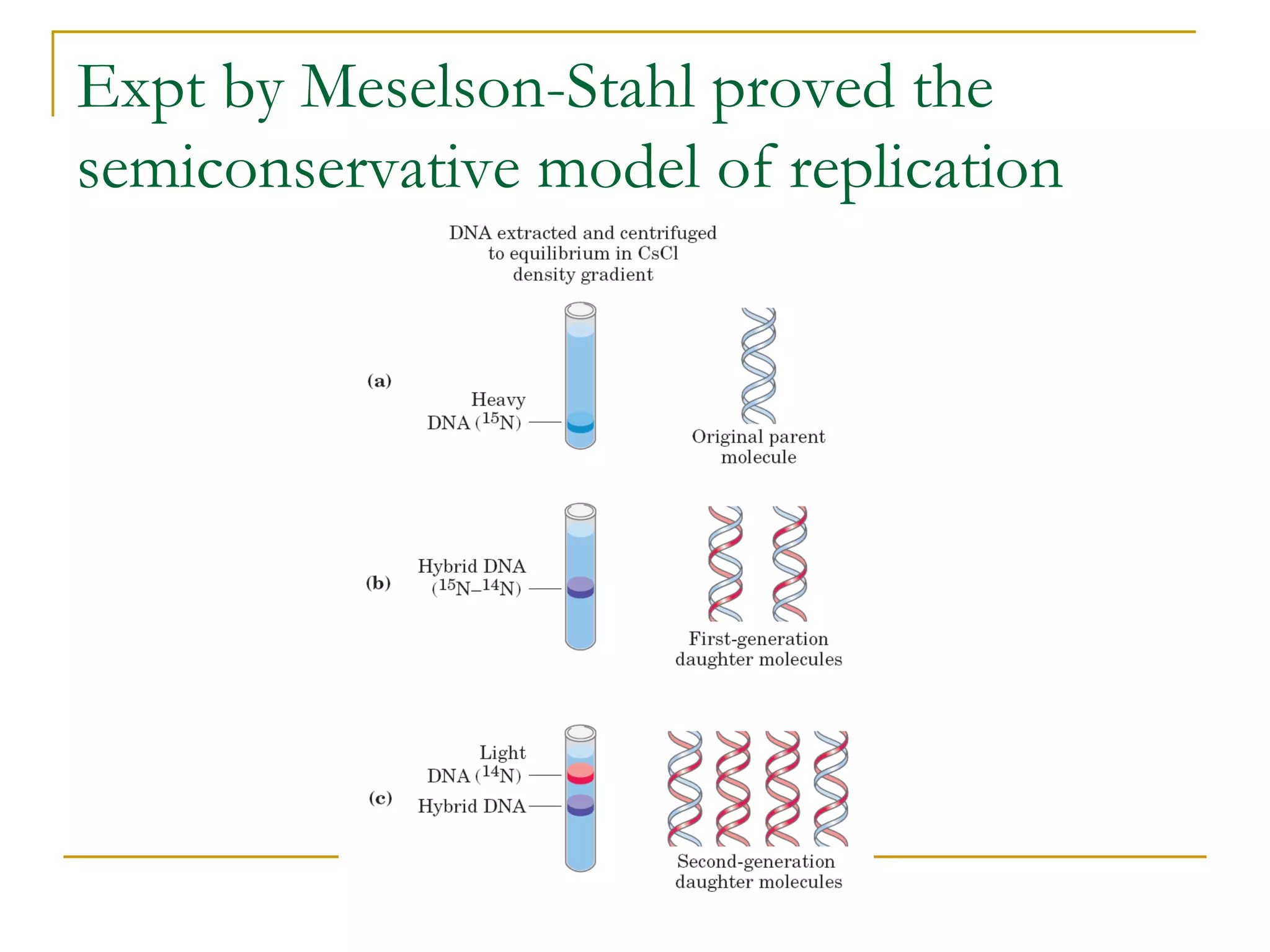
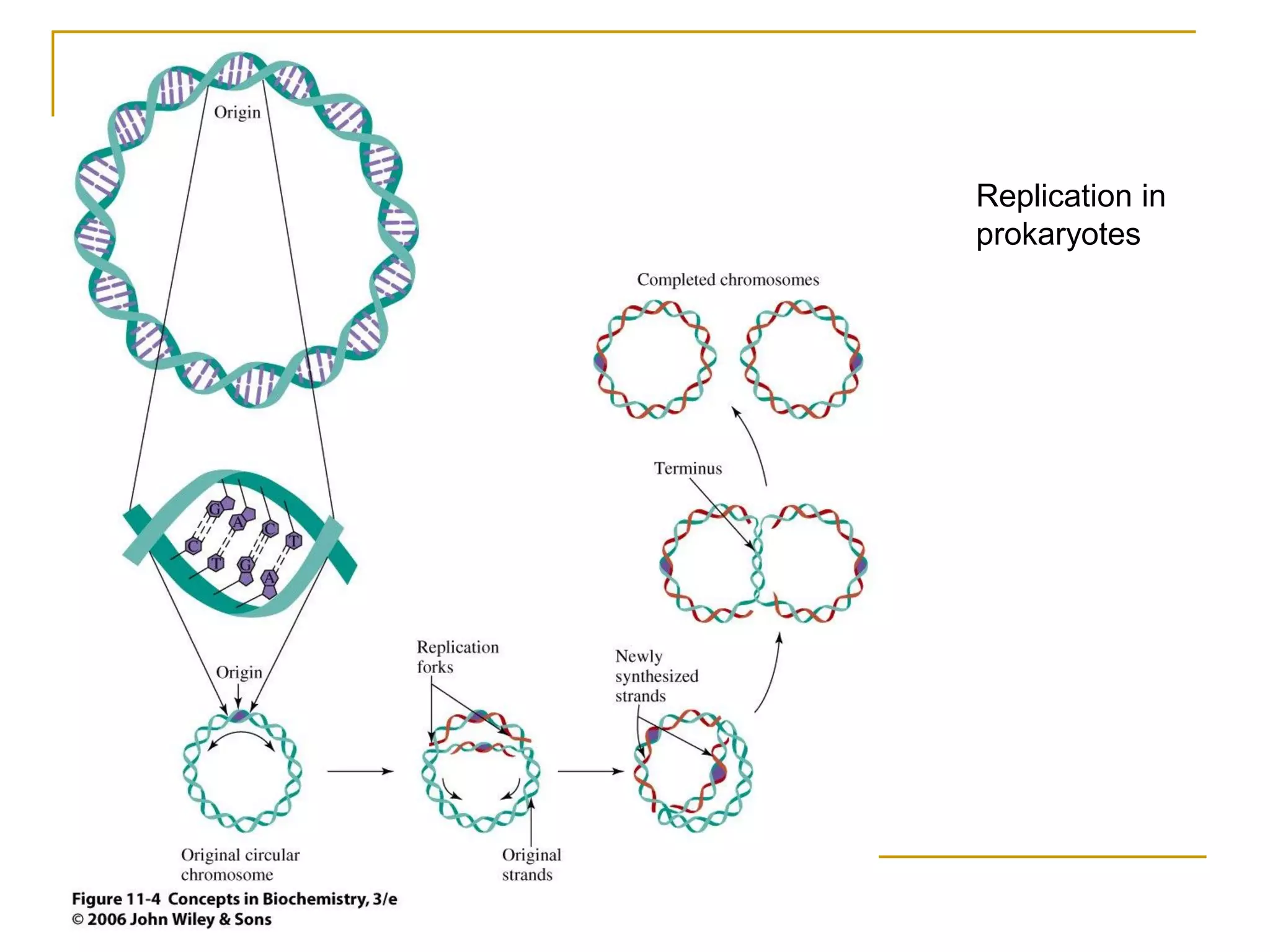
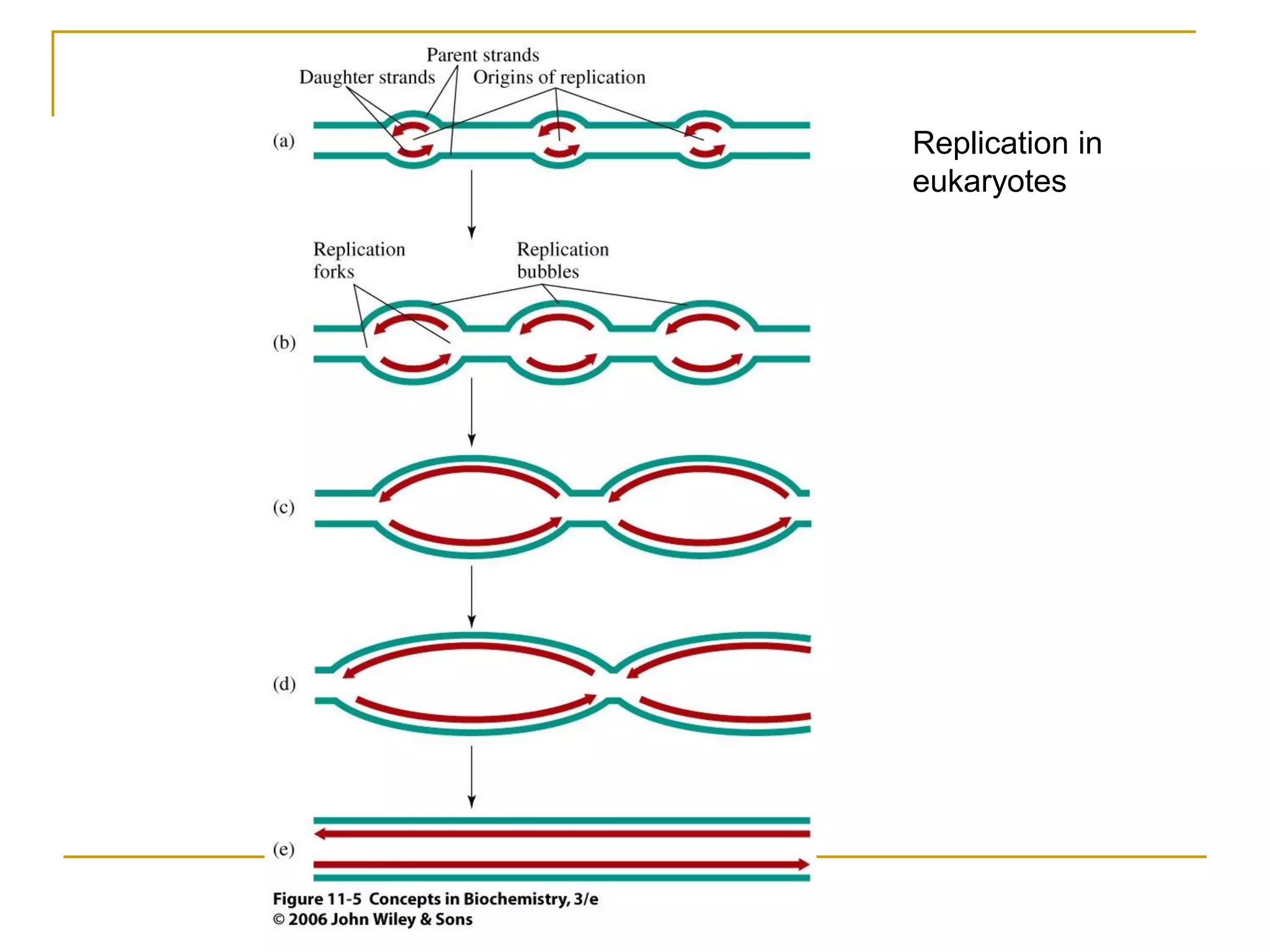
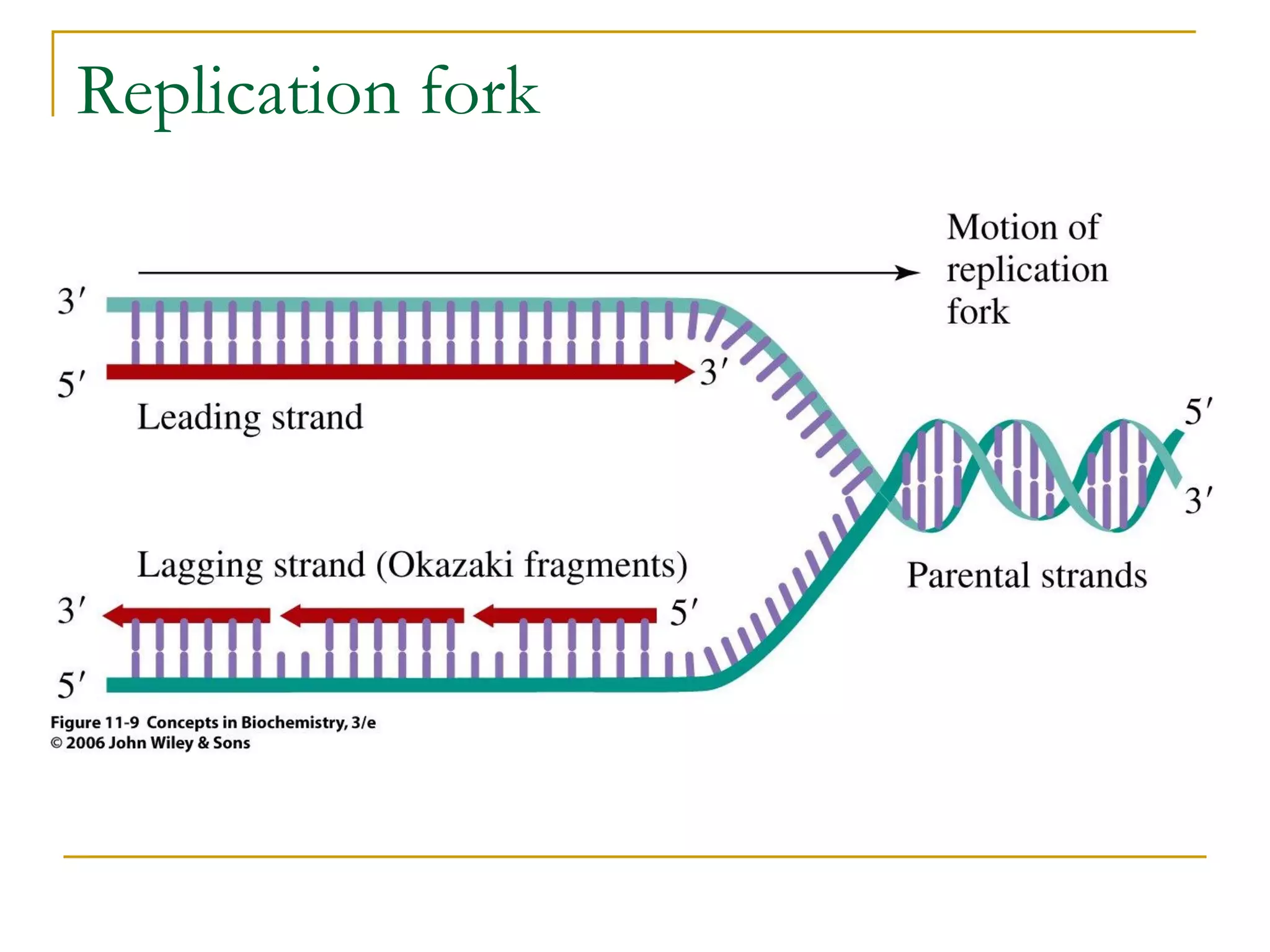
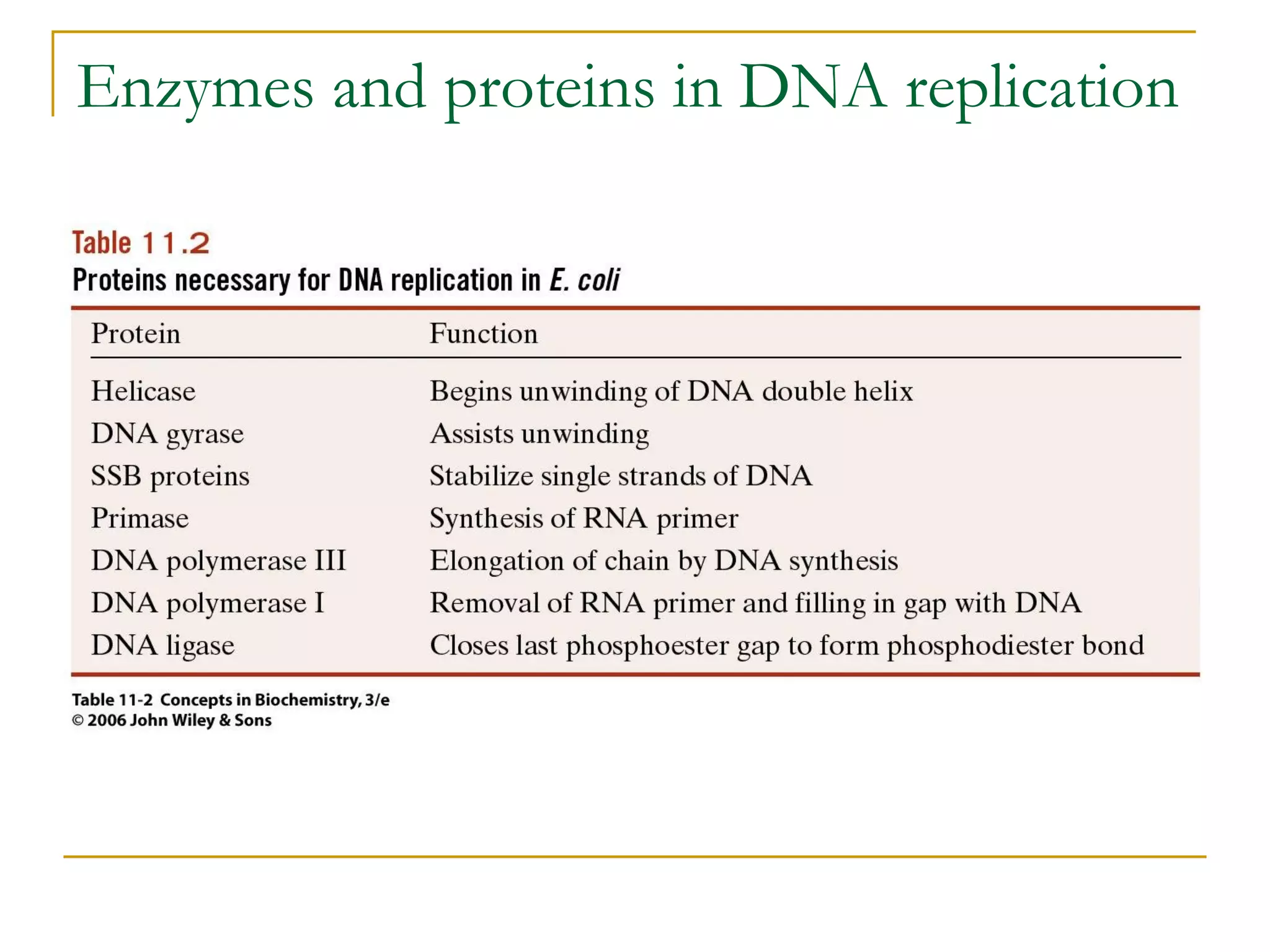
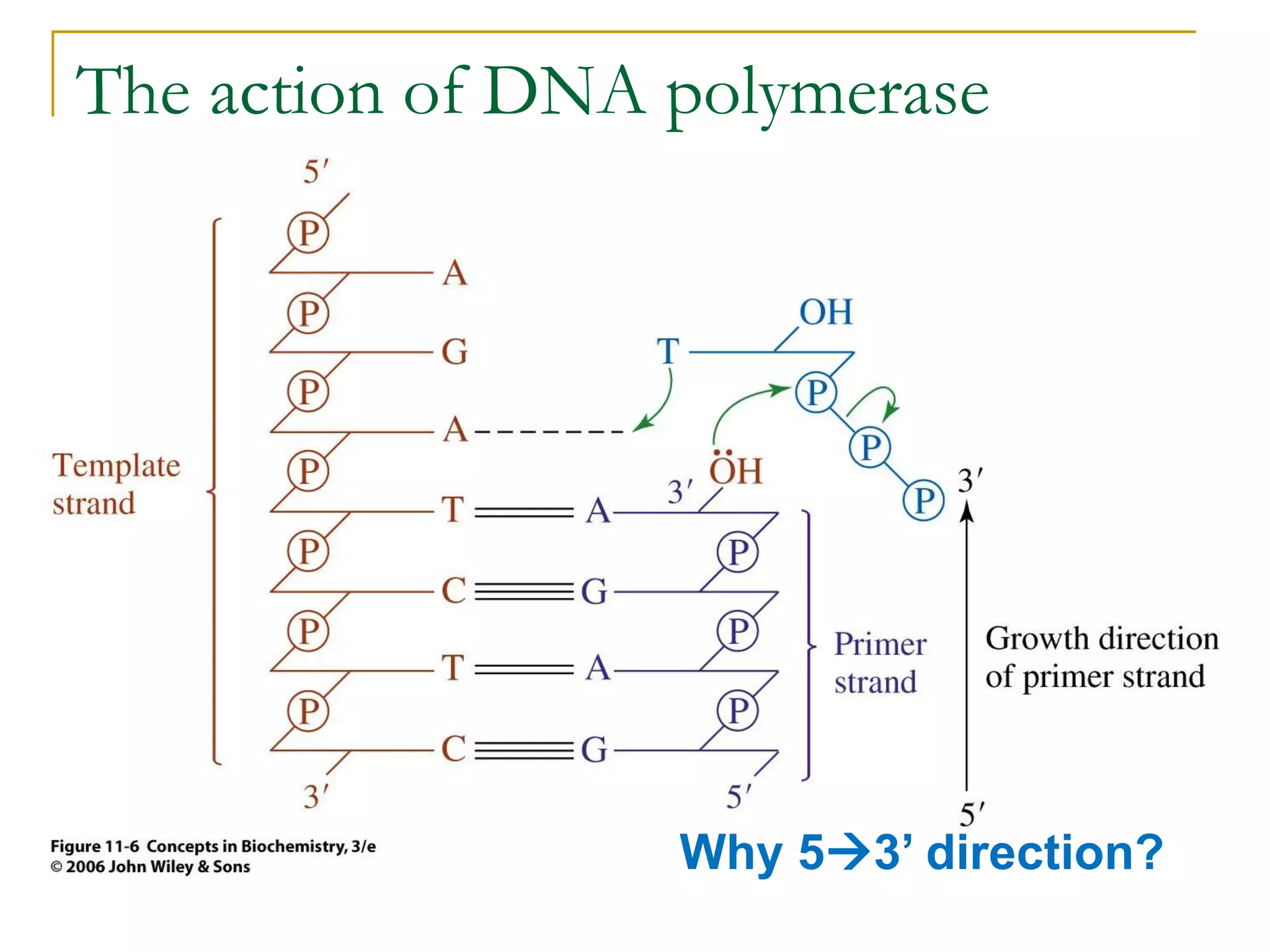
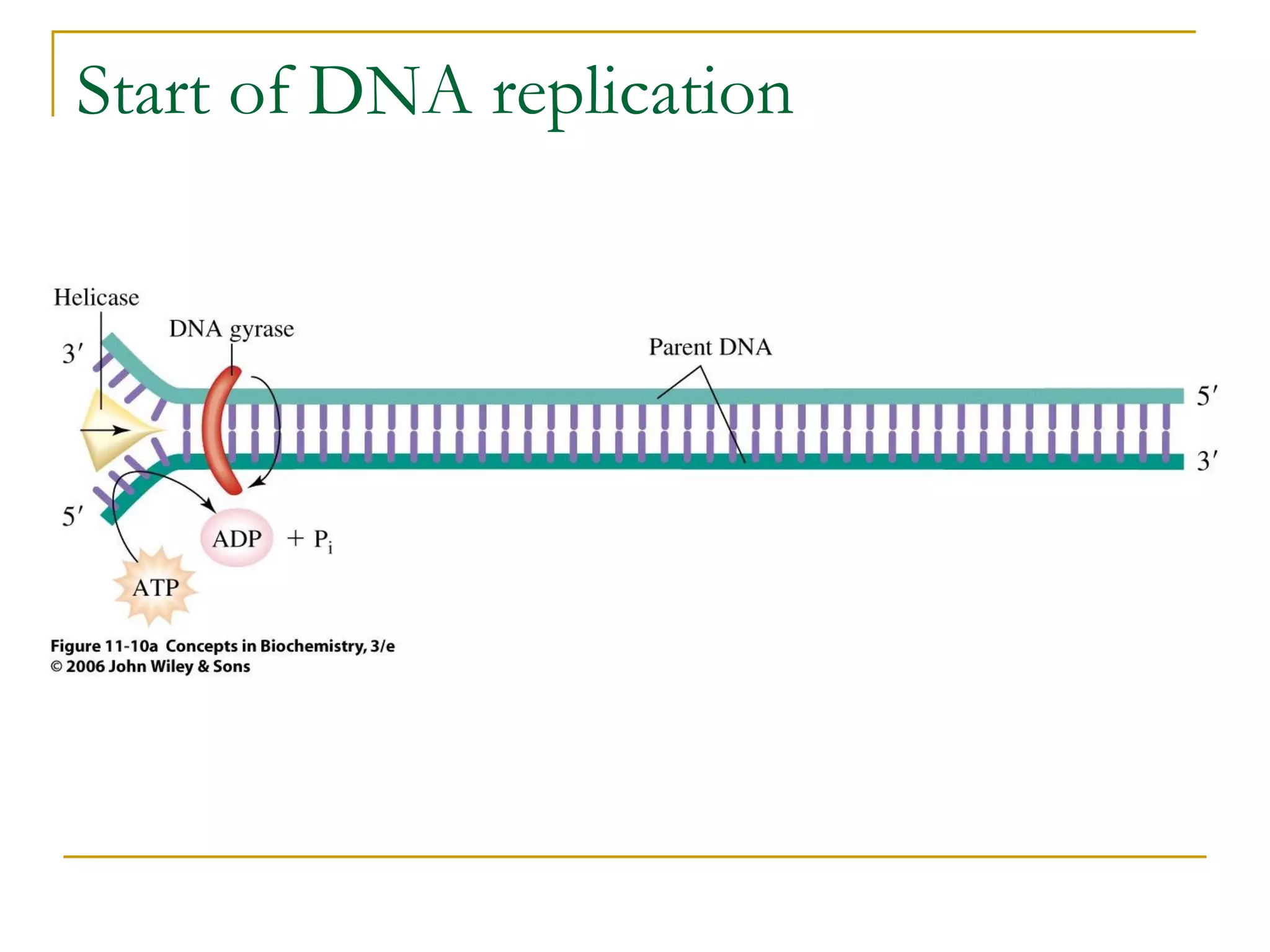
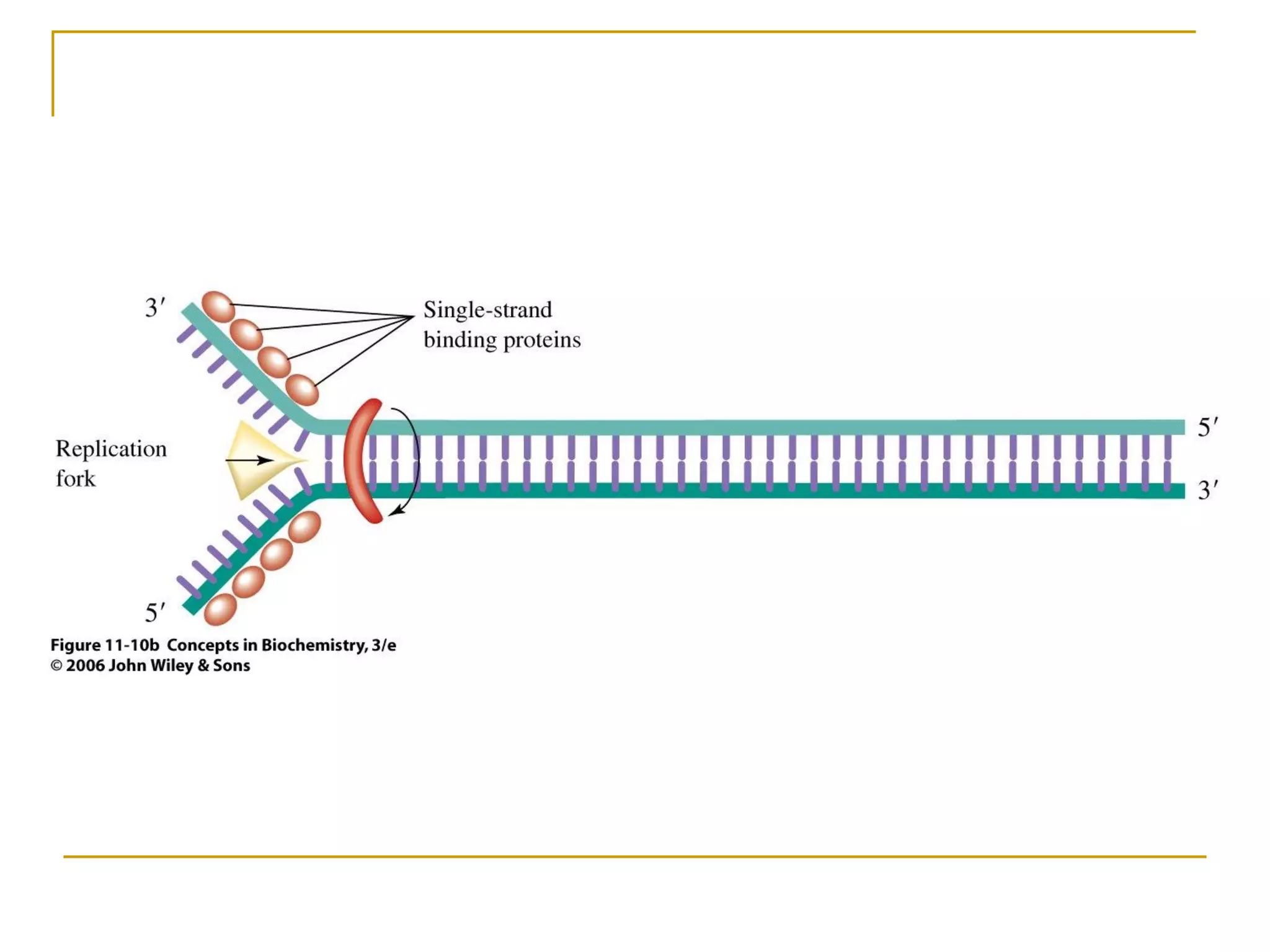
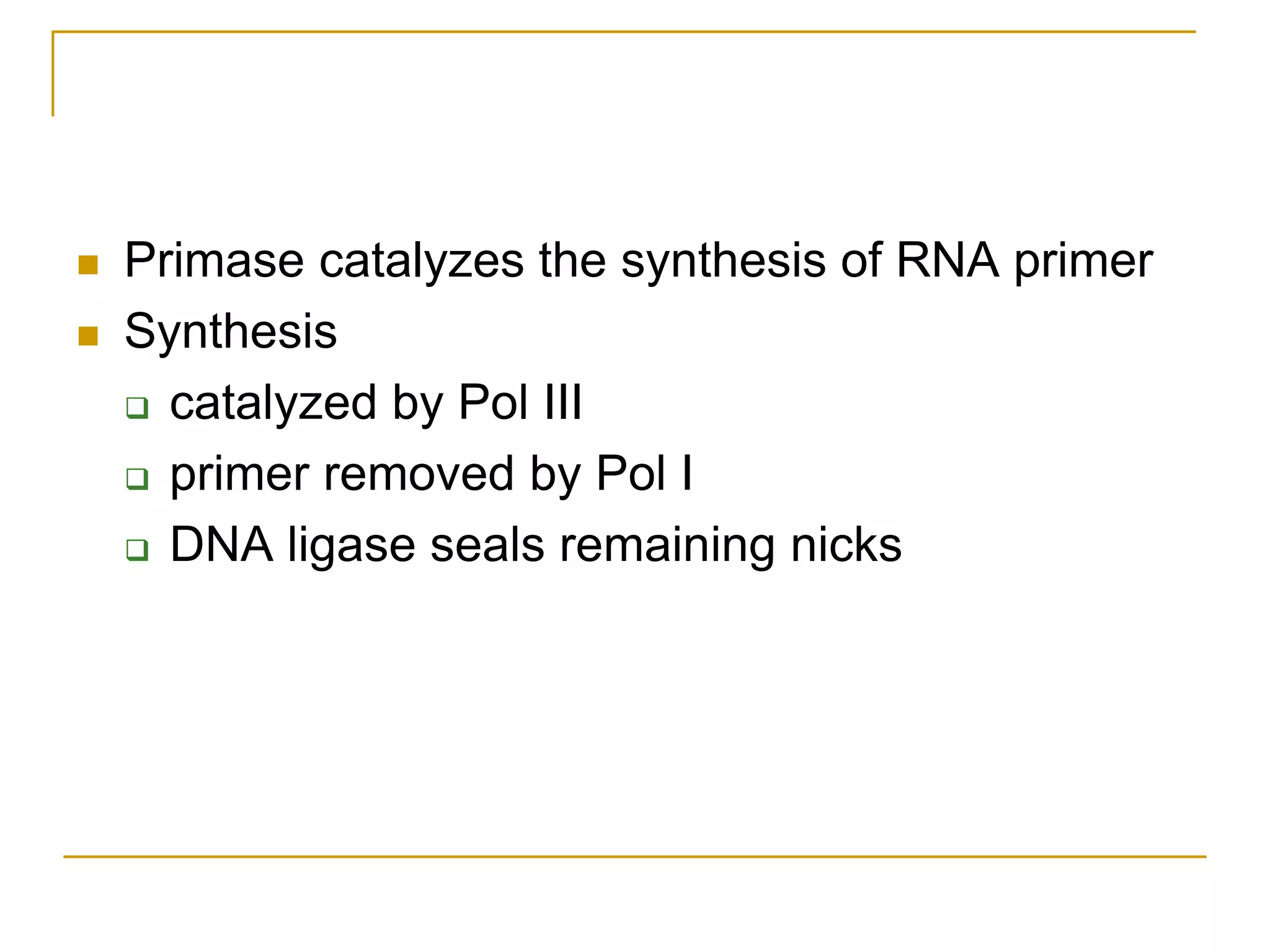
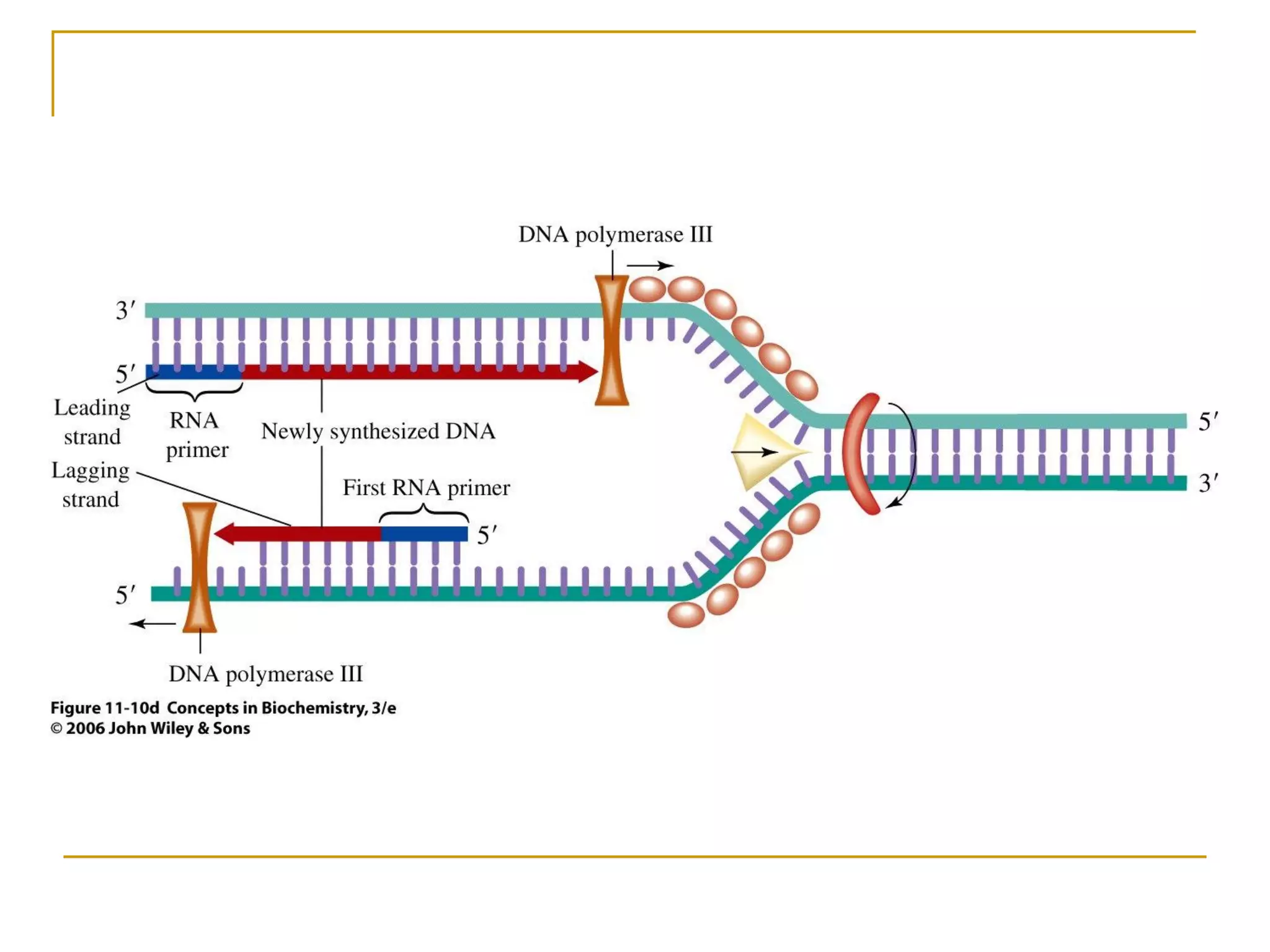
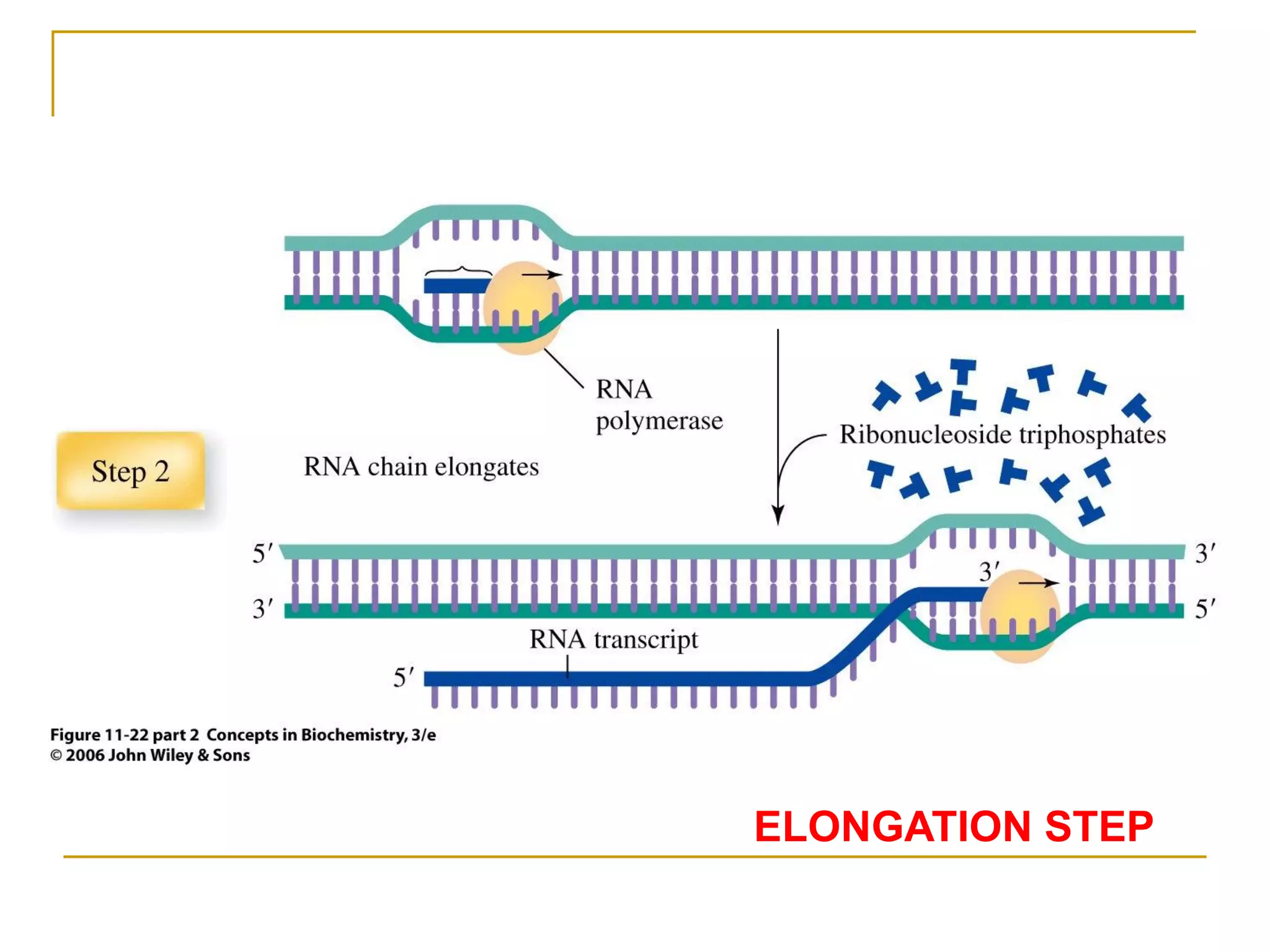
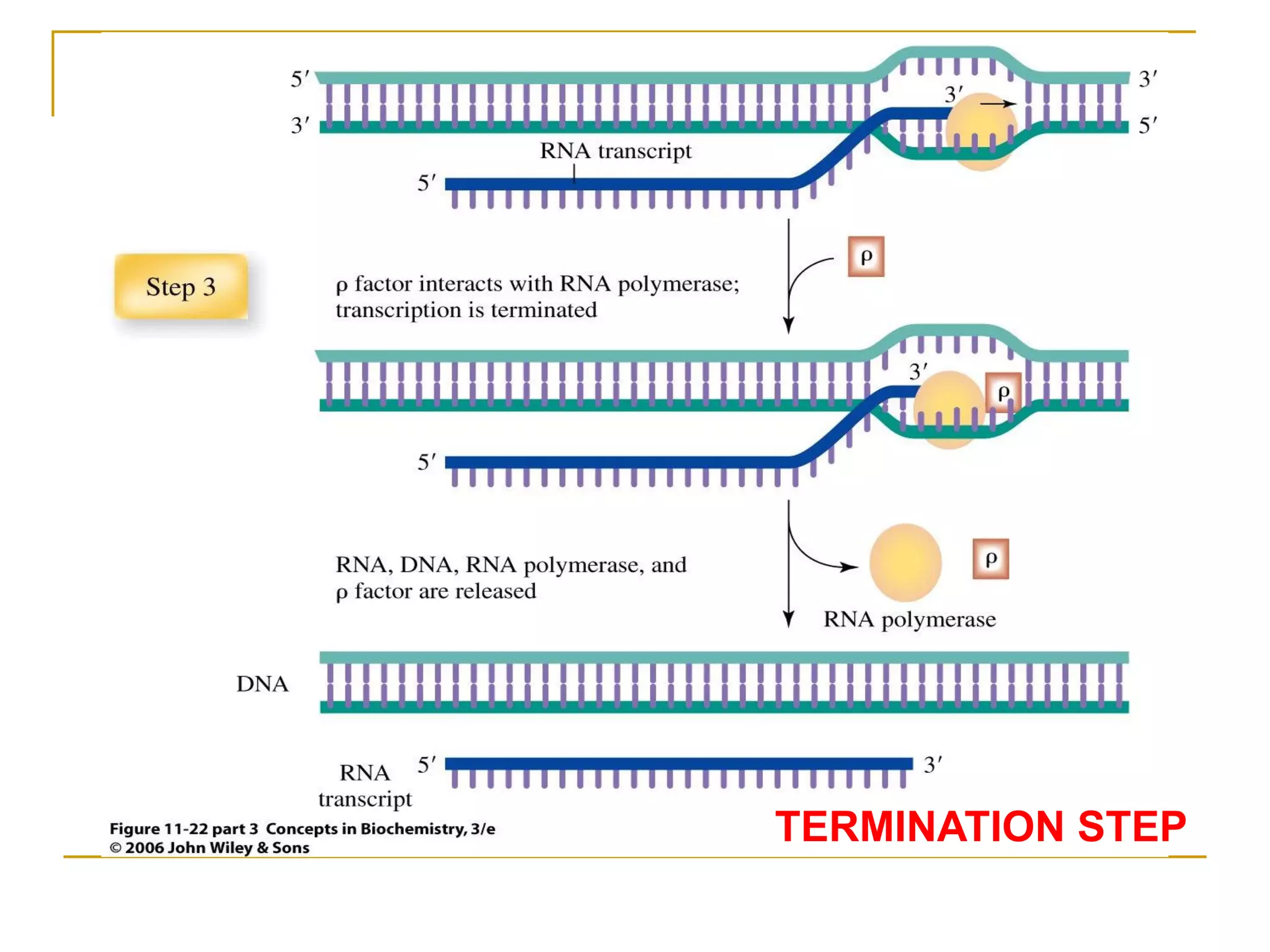
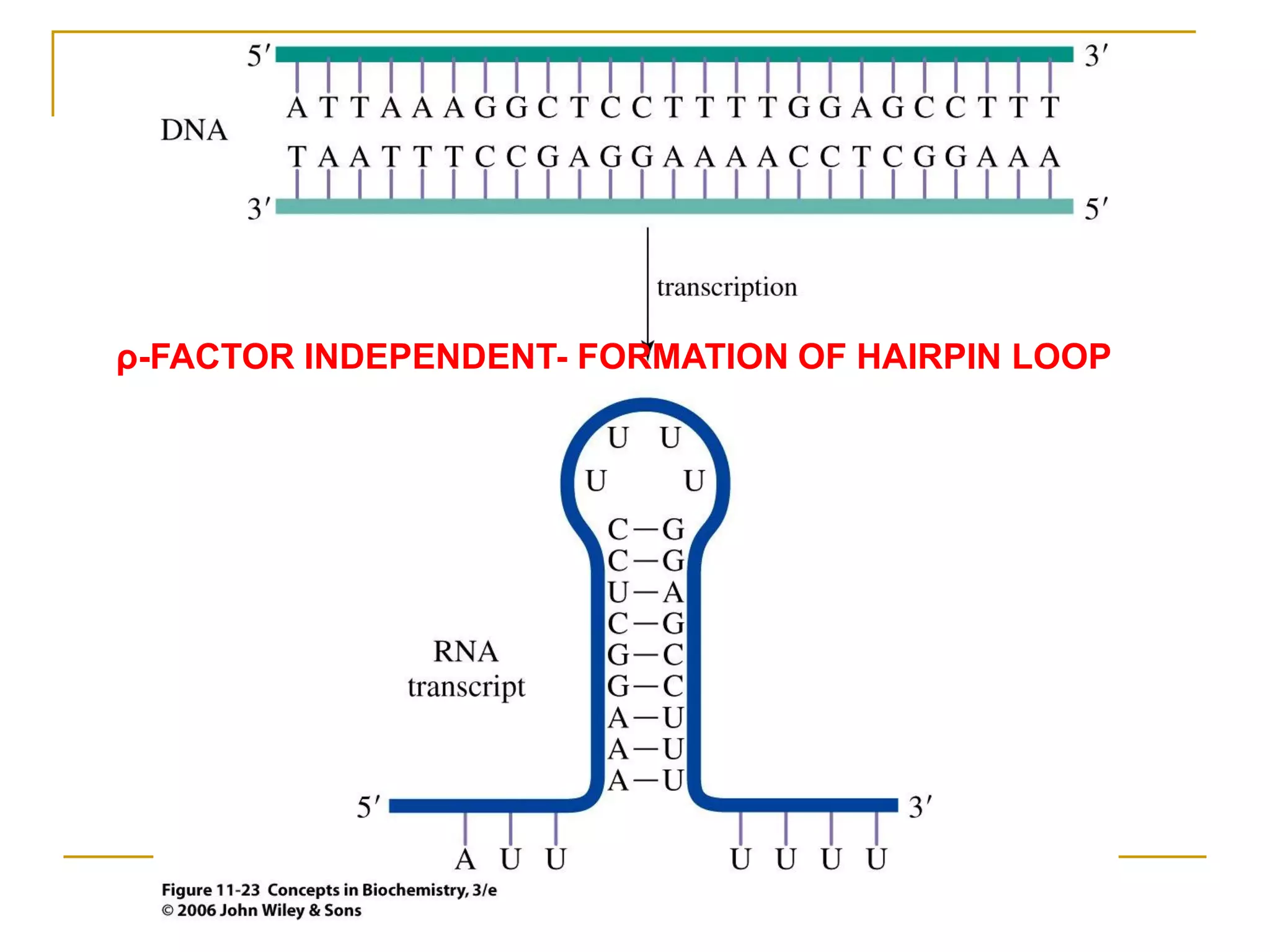
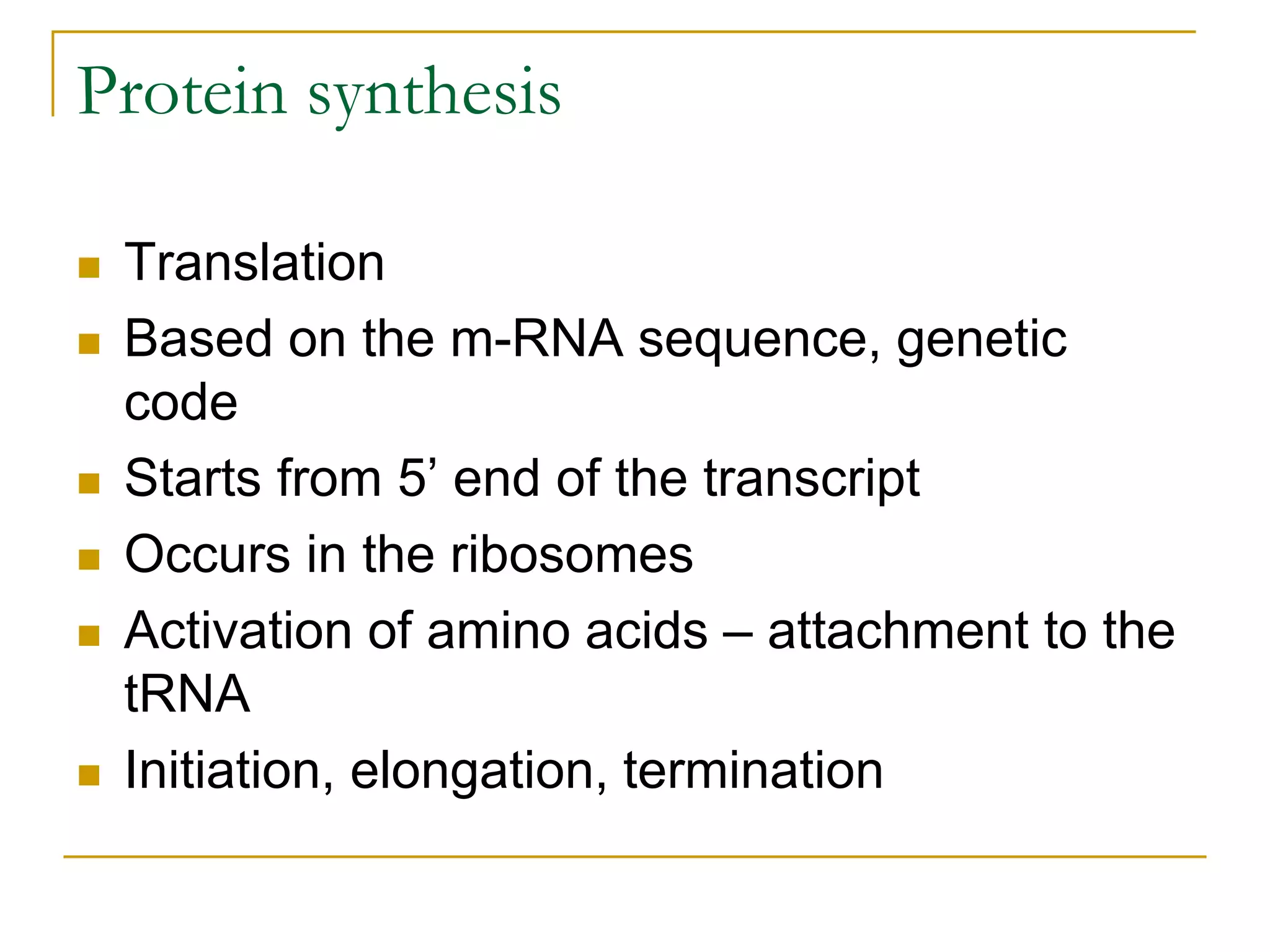
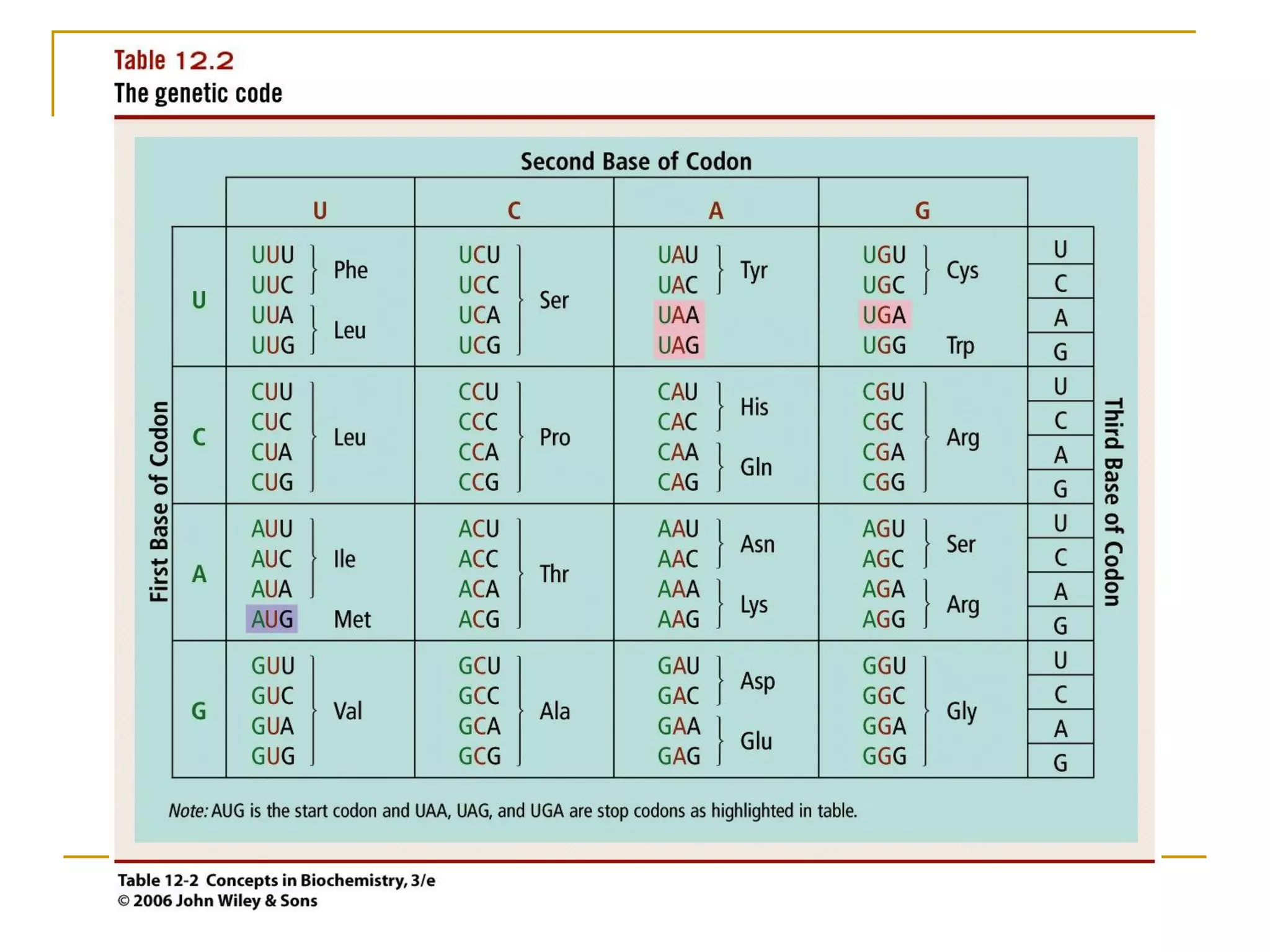
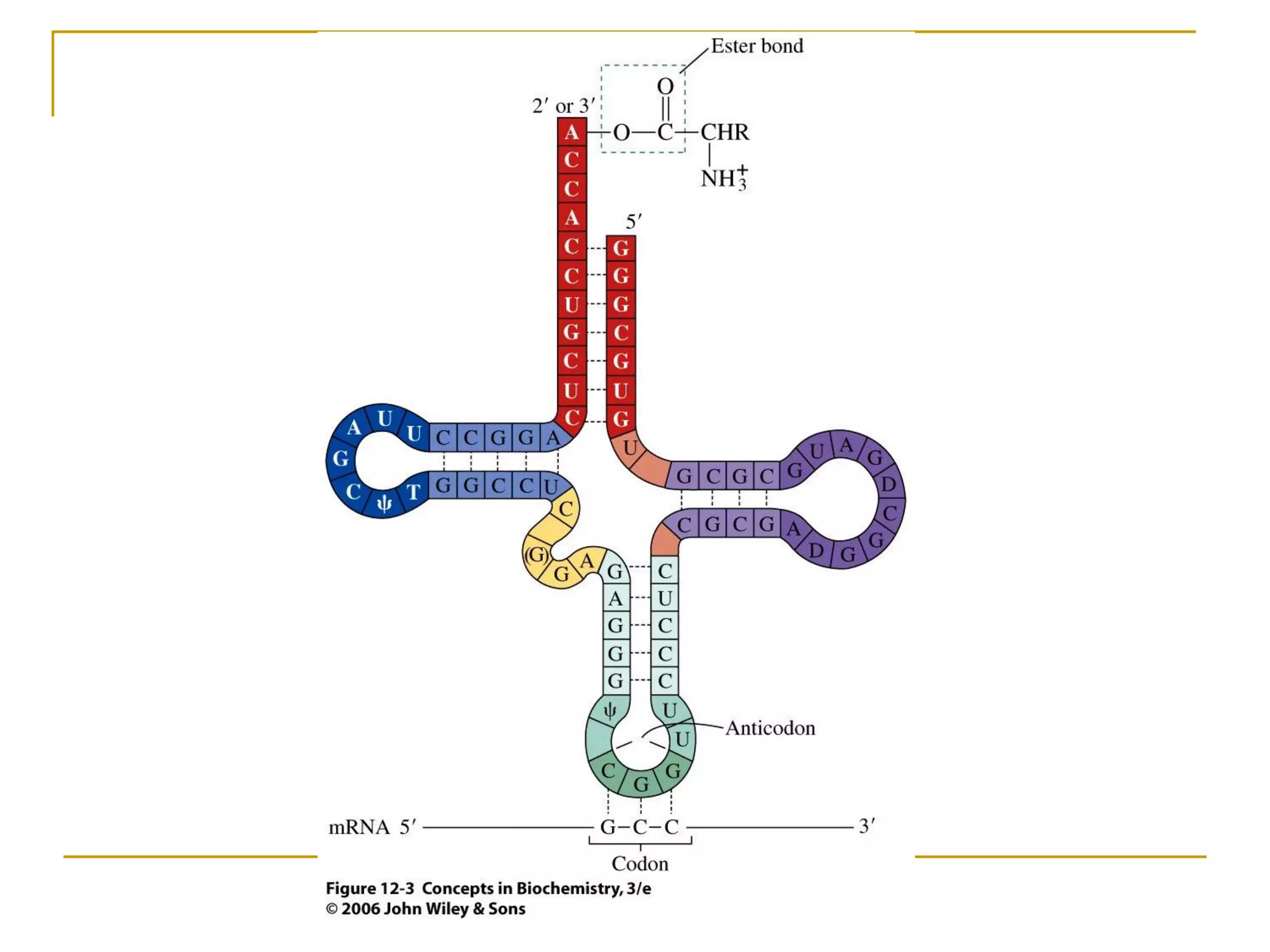
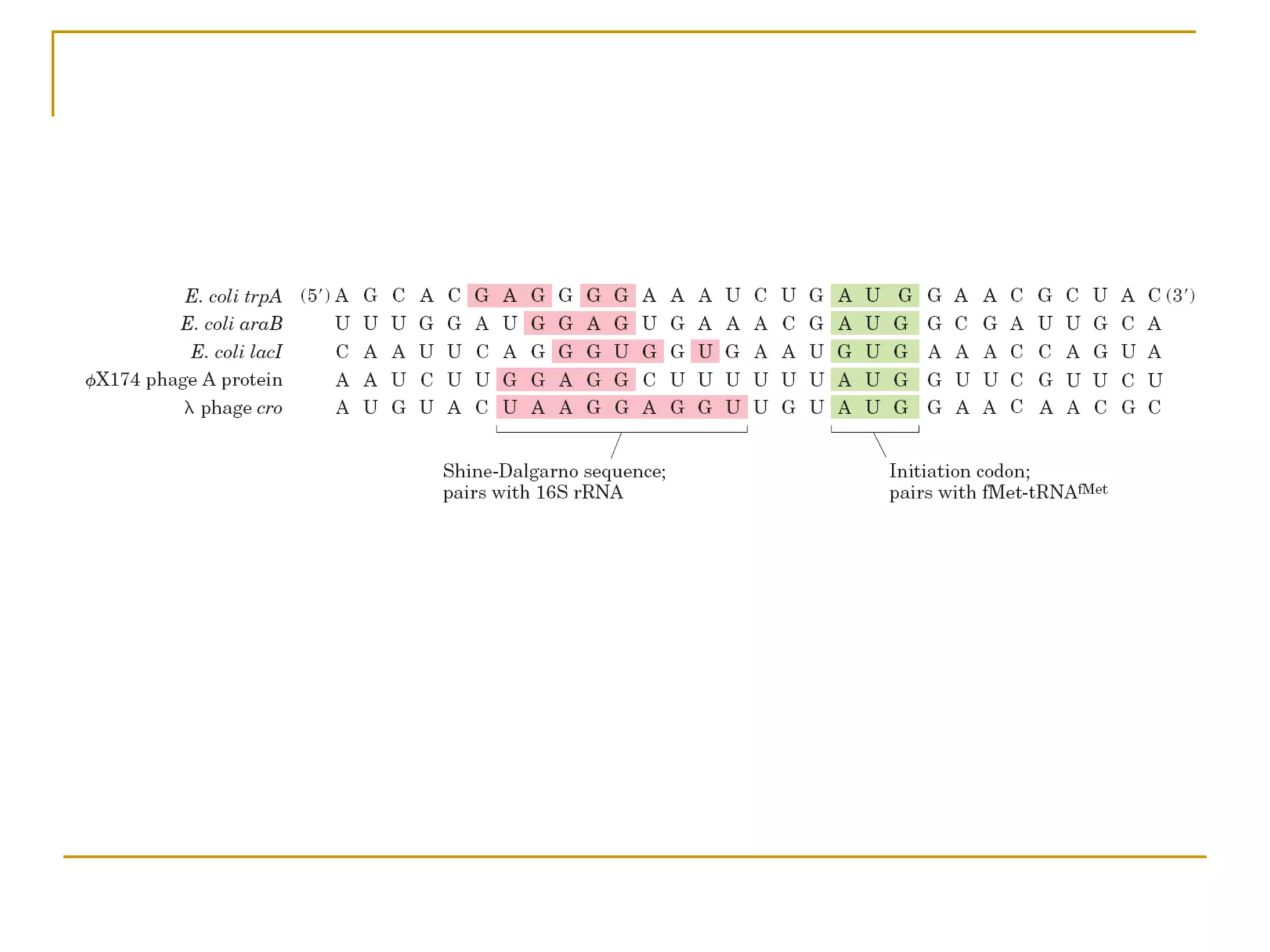
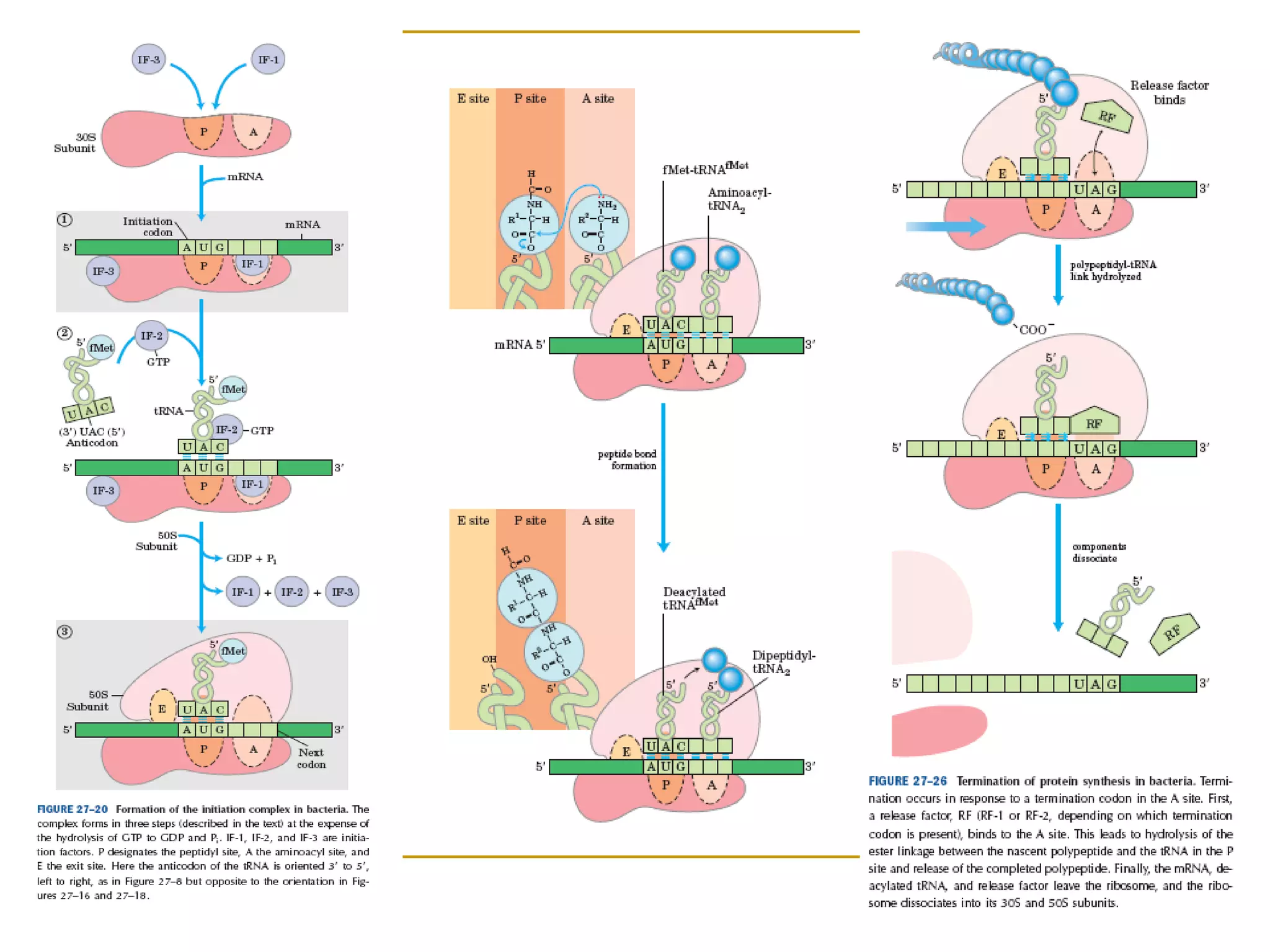
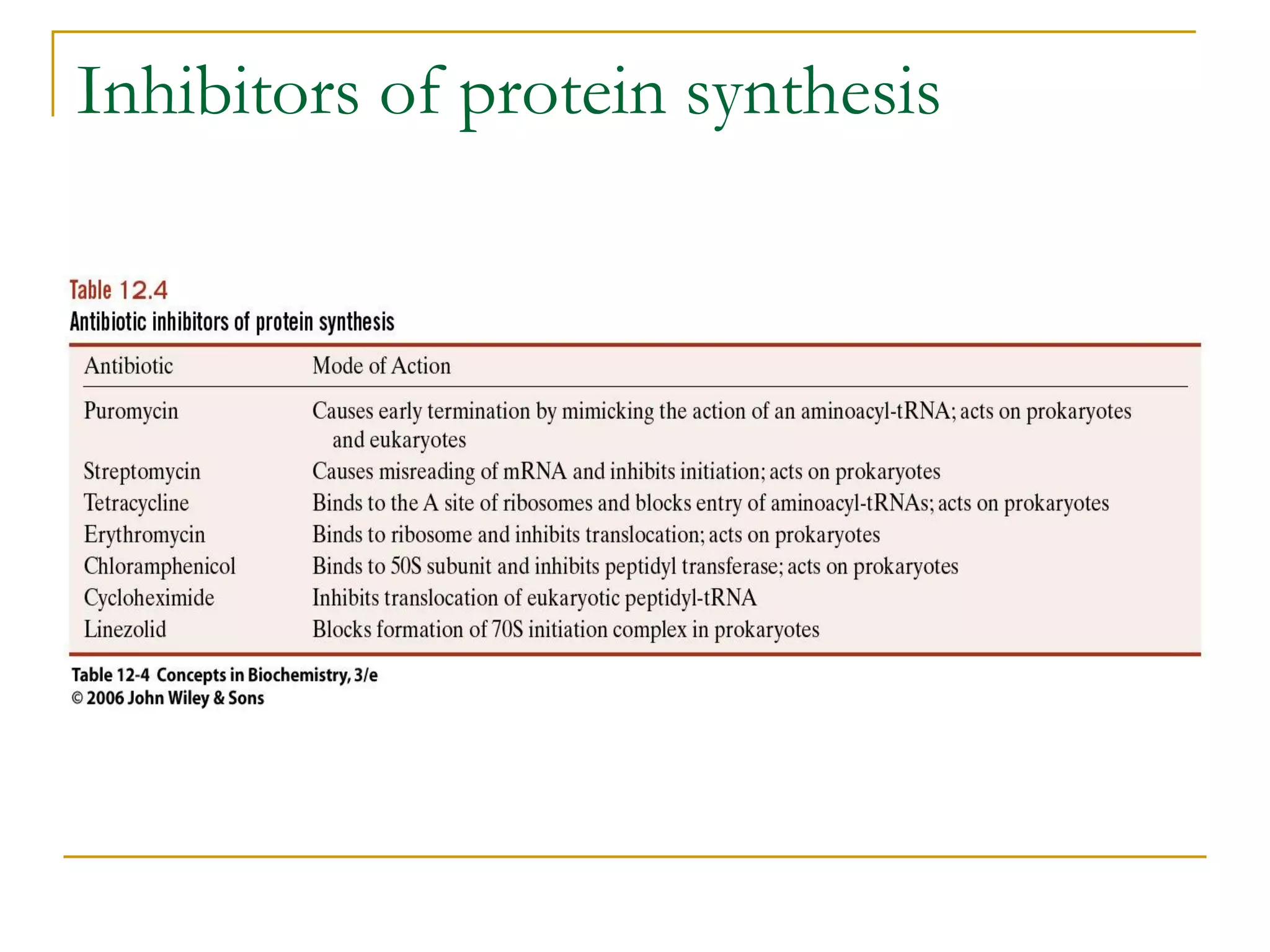
The document discusses the flow of genetic information from DNA to protein. It explains that DNA replication is semi-conservative and bidirectional, with the leading strand synthesized continuously and the lagging strand as Okazaki fragments. RNA synthesis involves transcription of DNA into RNA. Protein synthesis uses the genetic code to translate mRNA into amino acid sequences. The flow of genetic information and processes of replication, transcription, and translation are described.