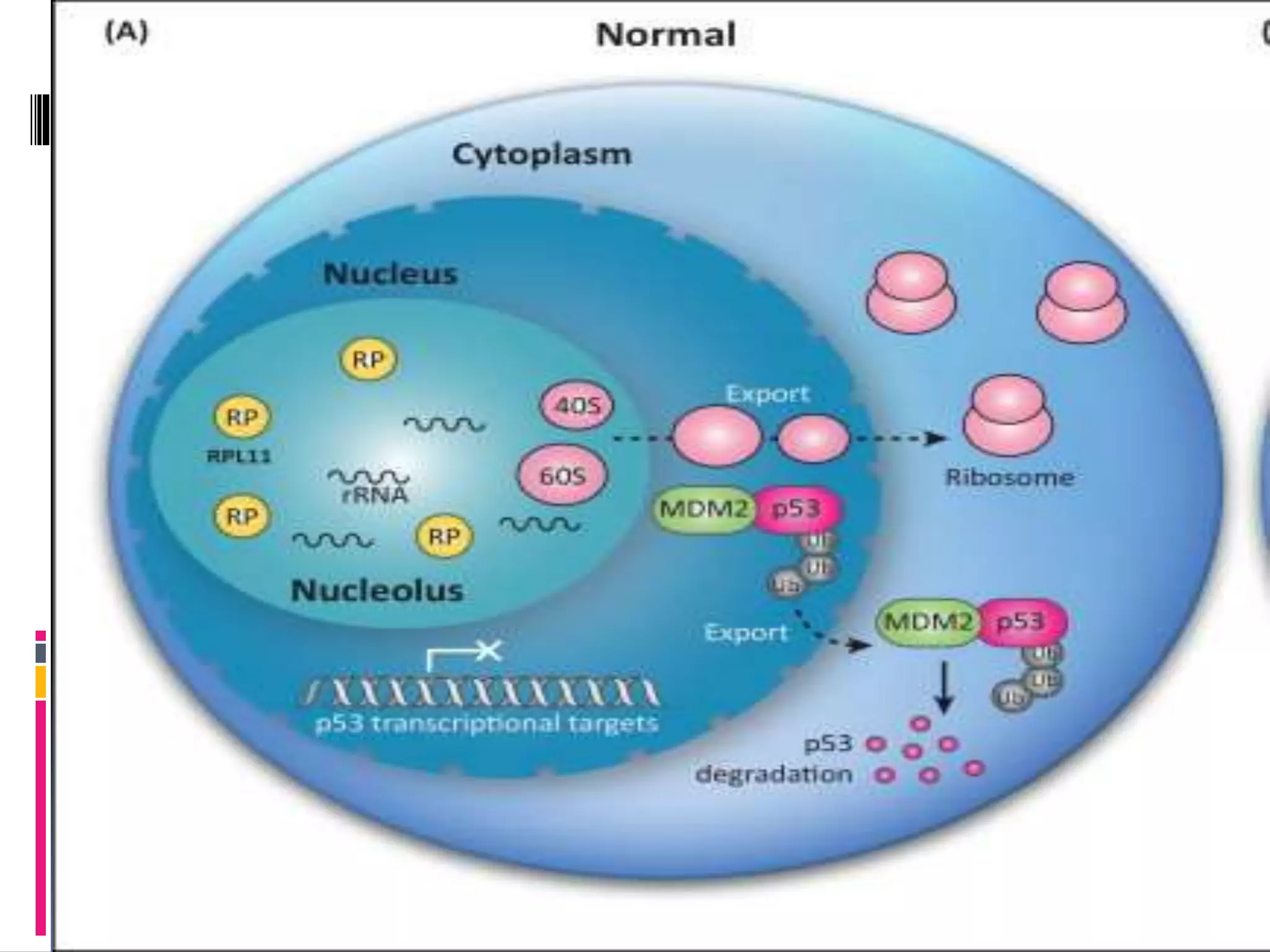
This document provides an overview of the nucleolus in plant cells. It discusses that the nucleolus is made up of fibrillar centers, dense fibrillar compartments, and granular centers, and its main function is ribosome biosynthesis. It also notes that the nucleolus is involved in other processes like RNA regulation, cell cycle progression, genome maintenance, and stress response. Specific proteins and pathways in each of these functions are outlined. Recent research topics discussed include the nucleolus's role in DNA repair, links to the proteasome, and effects of nucleolar protein deficiencies on plant growth and development.