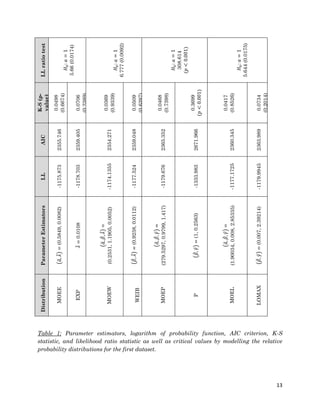
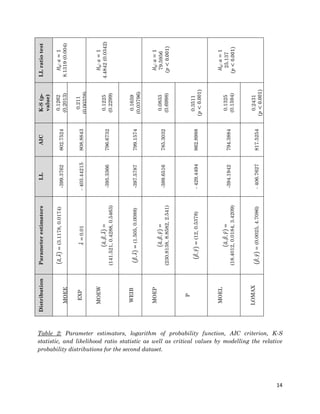
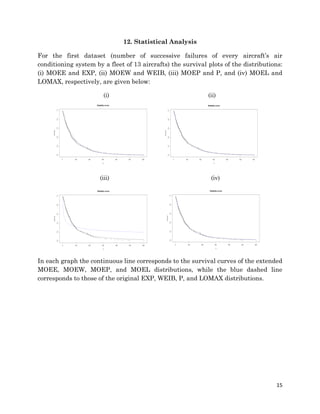
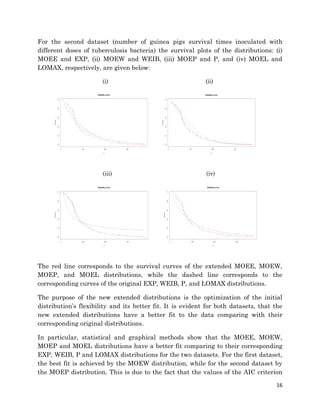
1. The document describes fitting several Marshall-Olkin distributions (MOEE, MOEW, MOEP, MOEL) as well as exponential, Weibull, Pareto and Lomax distributions to two real datasets using maximum likelihood estimation in R. For each distribution, it provides the cumulative distribution function, probability distribution function, likelihood function, and maximum likelihood estimates. It performs Kolmogorov-Smirnov goodness of fit tests to compare the fitted distributions to the datasets.
2. Functions for the cumulative distribution function, probability distribution function, and likelihood function are provided for each of the MOEE, MOEW, MOEP, MOEL and MOELFR distributions. Maximum likelihood estimation and Kolmogorov-Sm