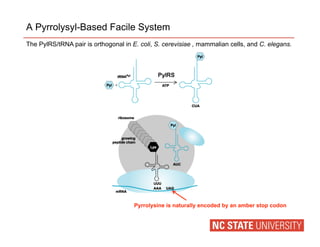
This document discusses incorporating unnatural amino acids into proteins to expand their functions. It describes how pyrrolysine is used as an orthogonal system to incorporate unnatural amino acids specified by an amber stop codon. Various unnatural amino acids containing alkene and norbornene functional groups were successfully incorporated into GFP and other proteins in E. coli and mammalian cells. These modified proteins could then be site-specifically labeled using bioorthogonal reactions like the thiol-ene and Diels-Alder reactions. This allows proteins to be labeled and studied in live cells with new functions.