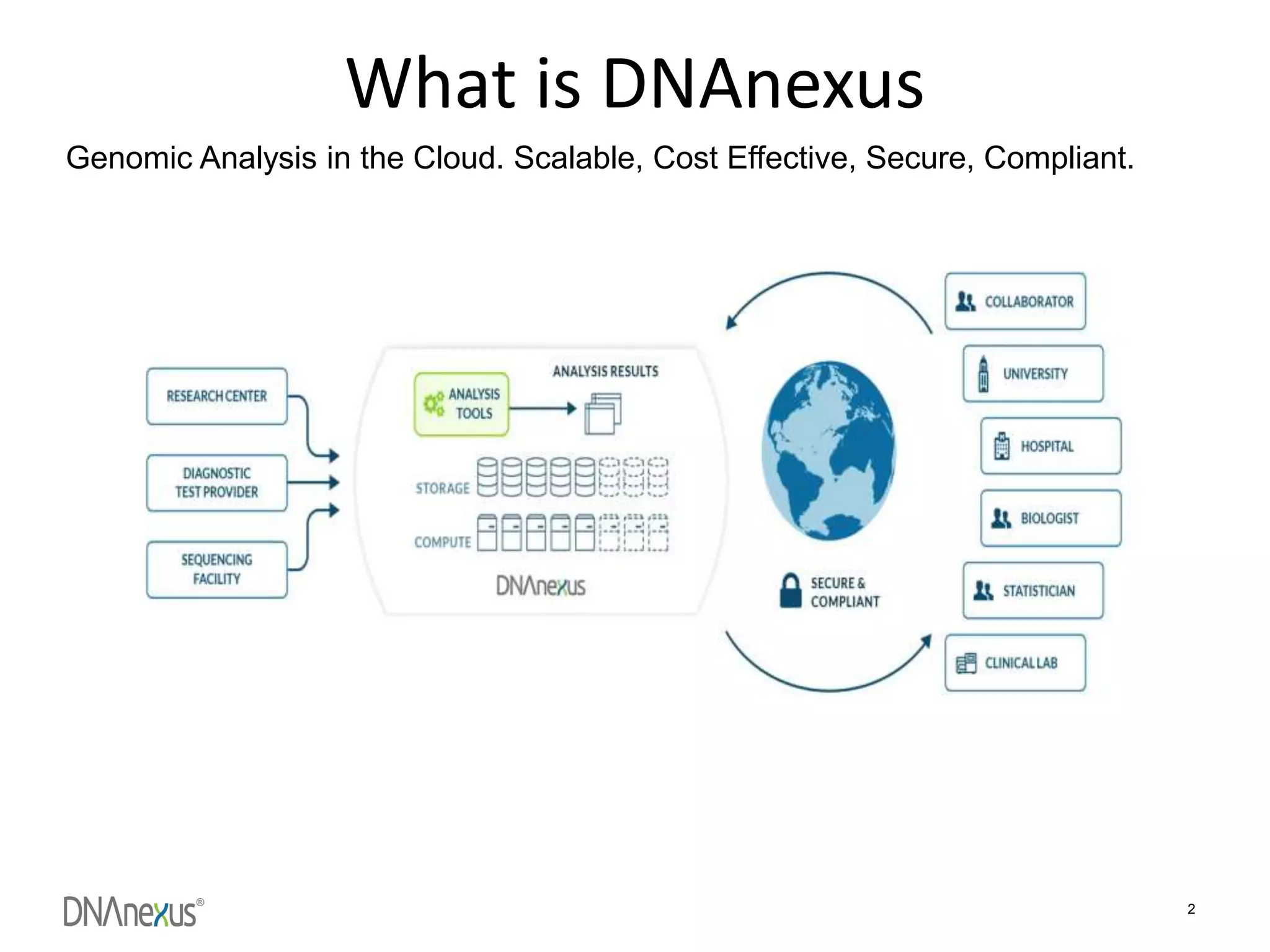
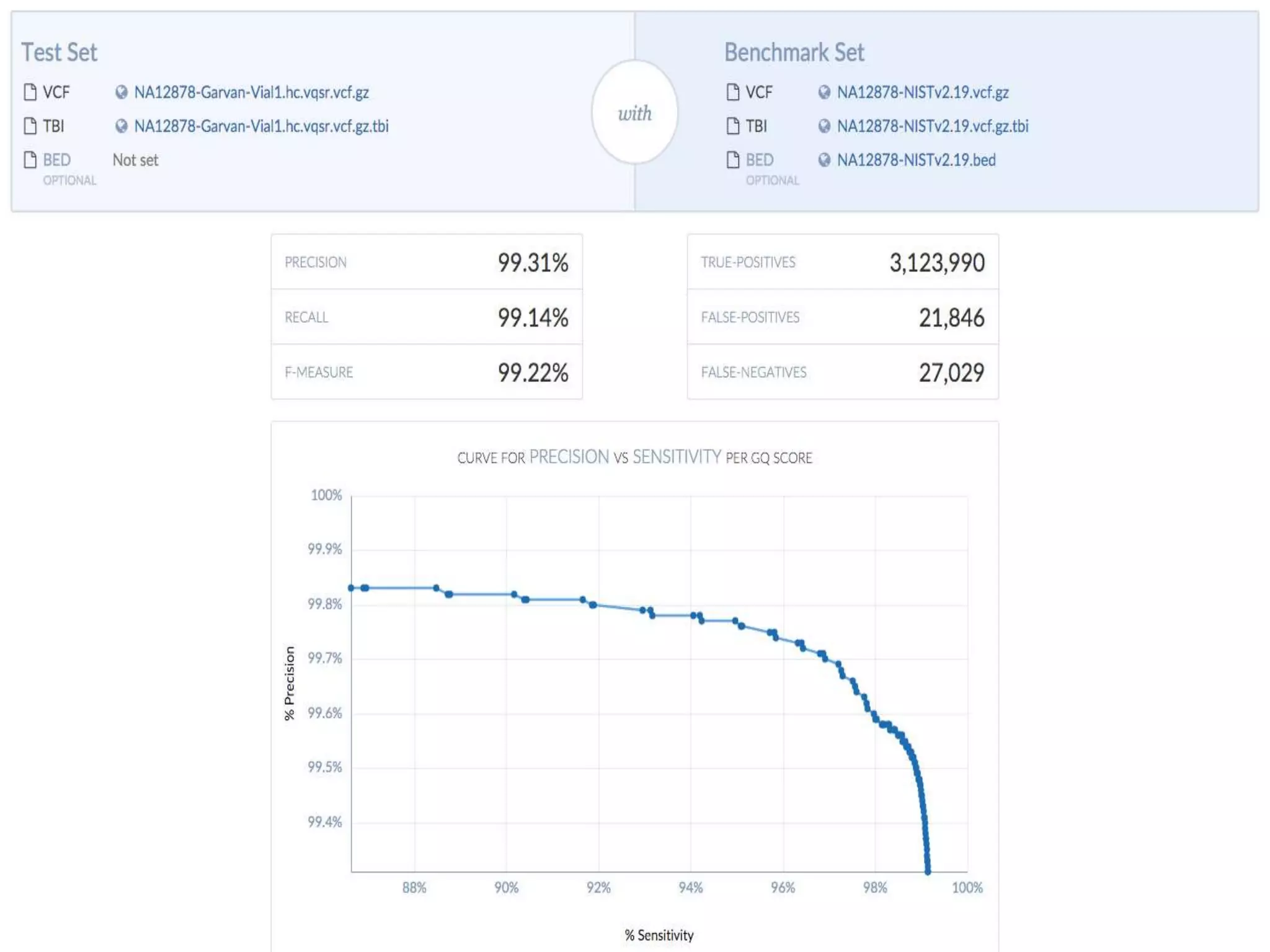
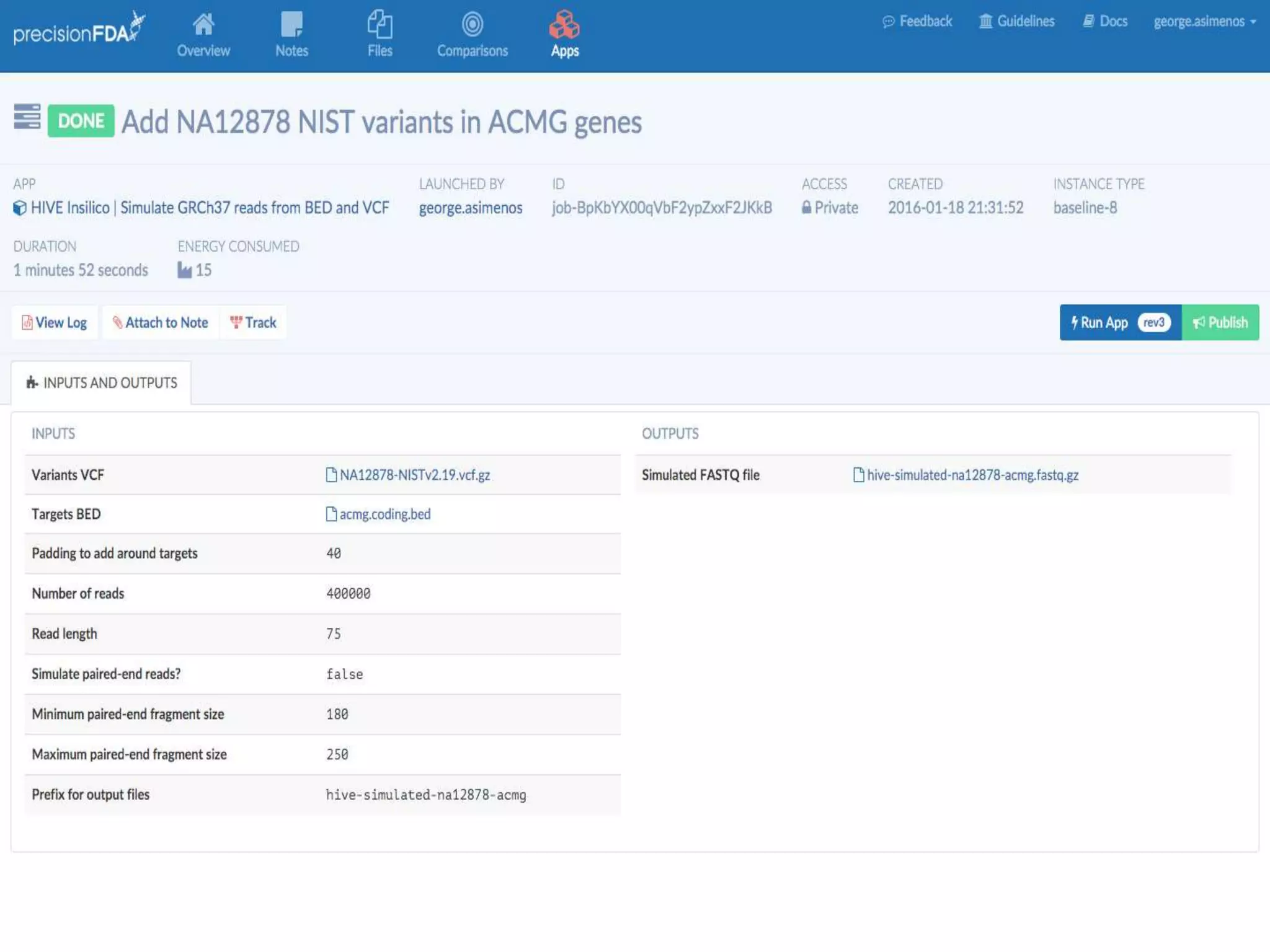
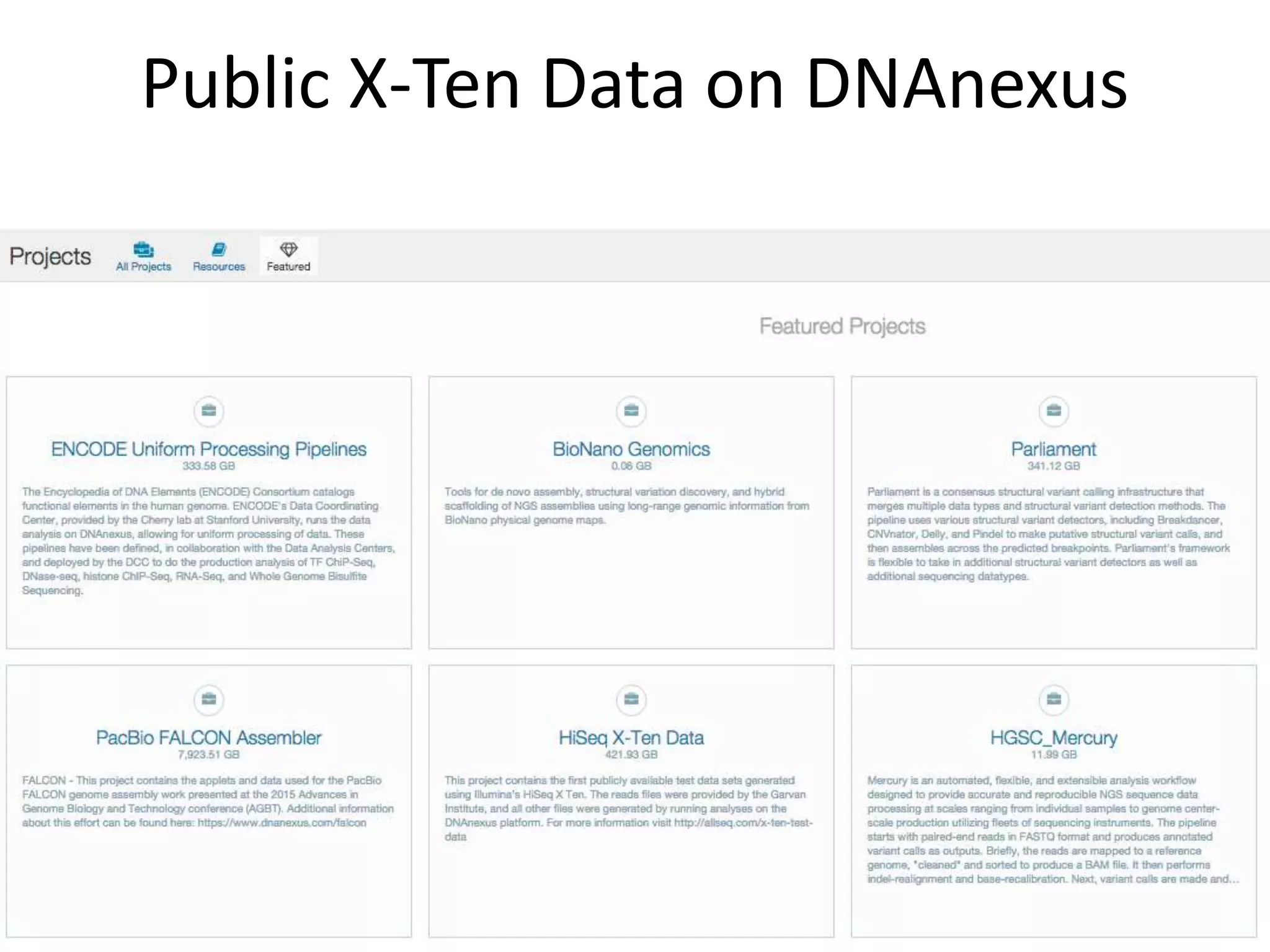
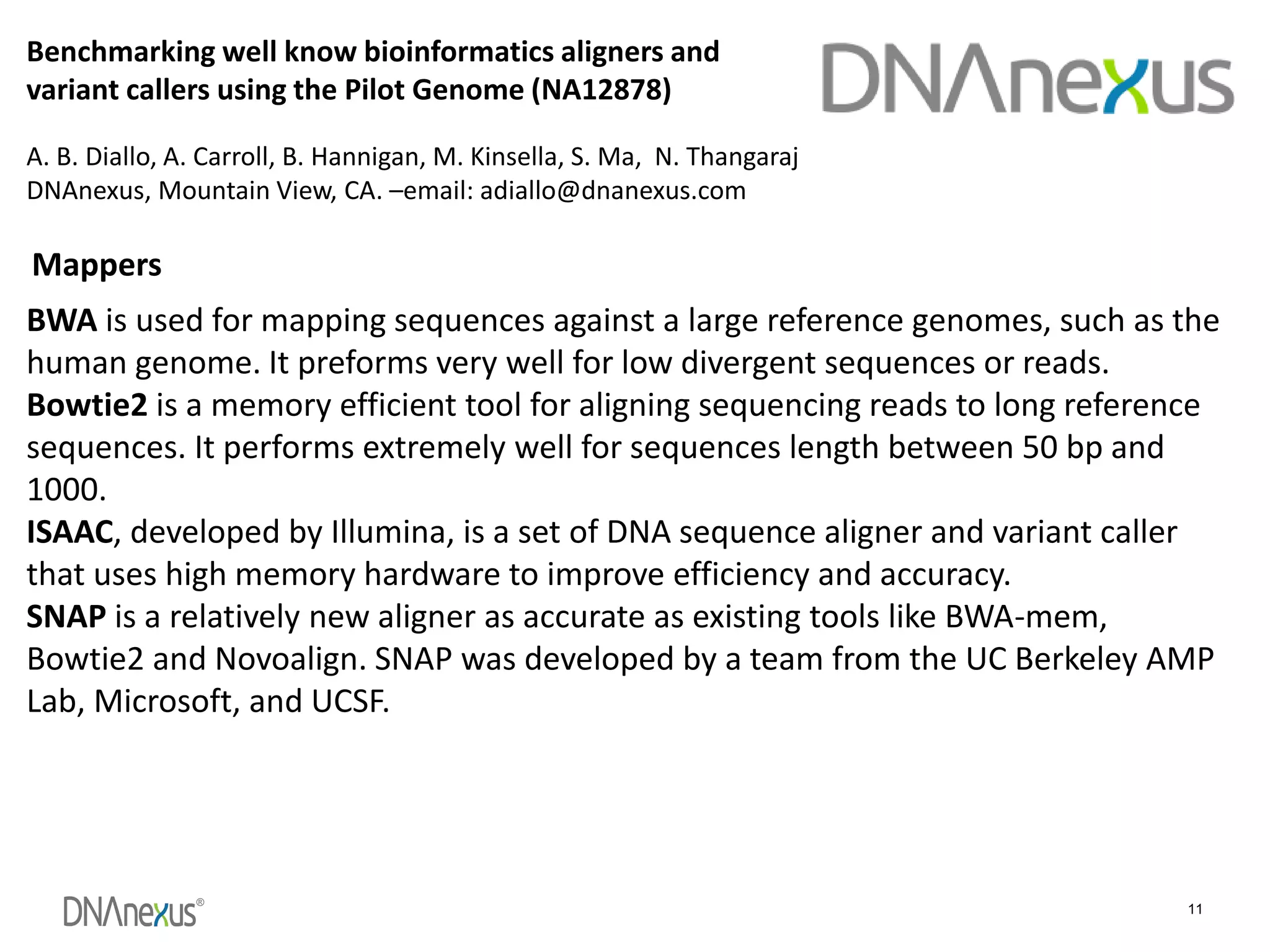
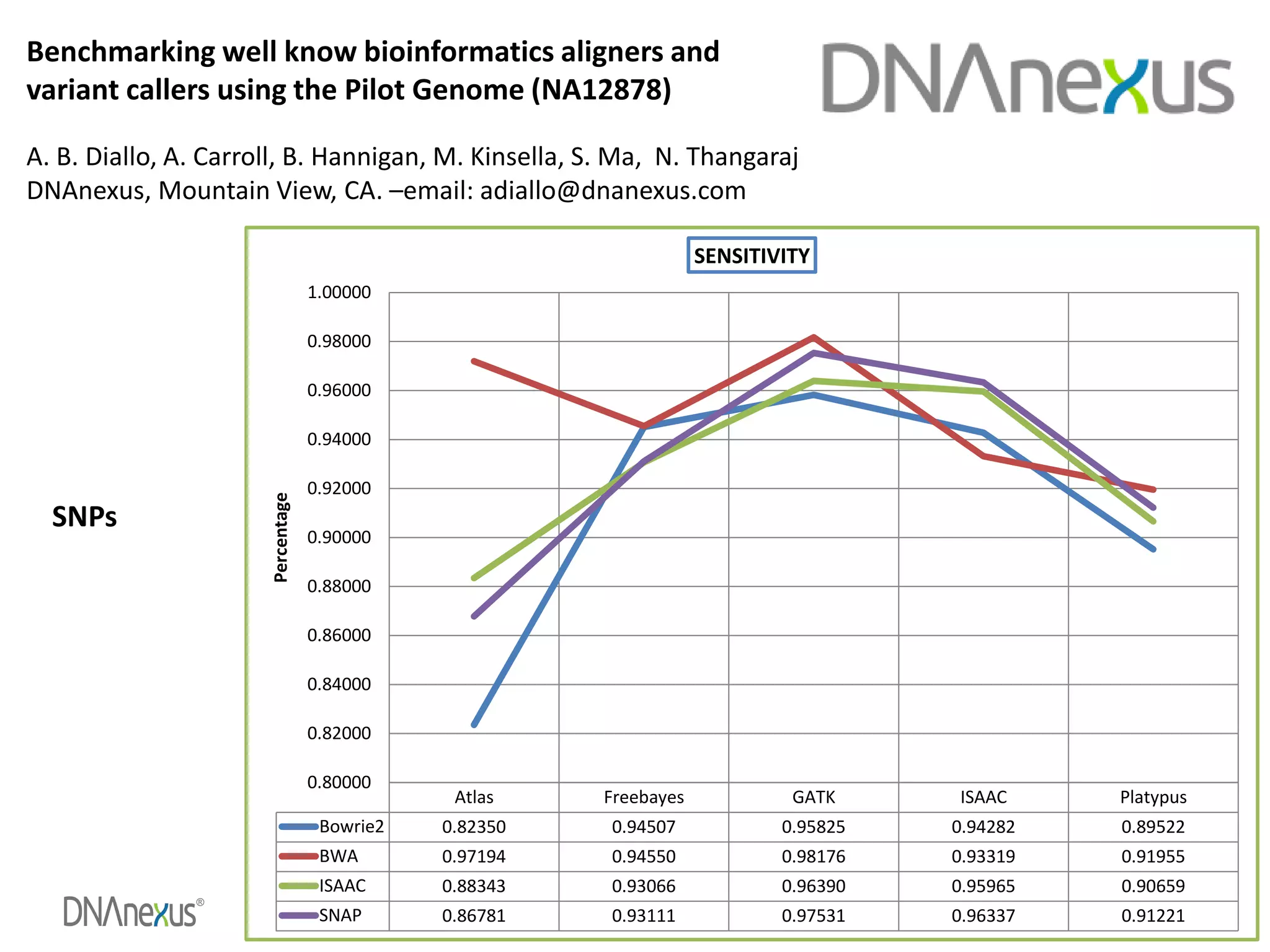
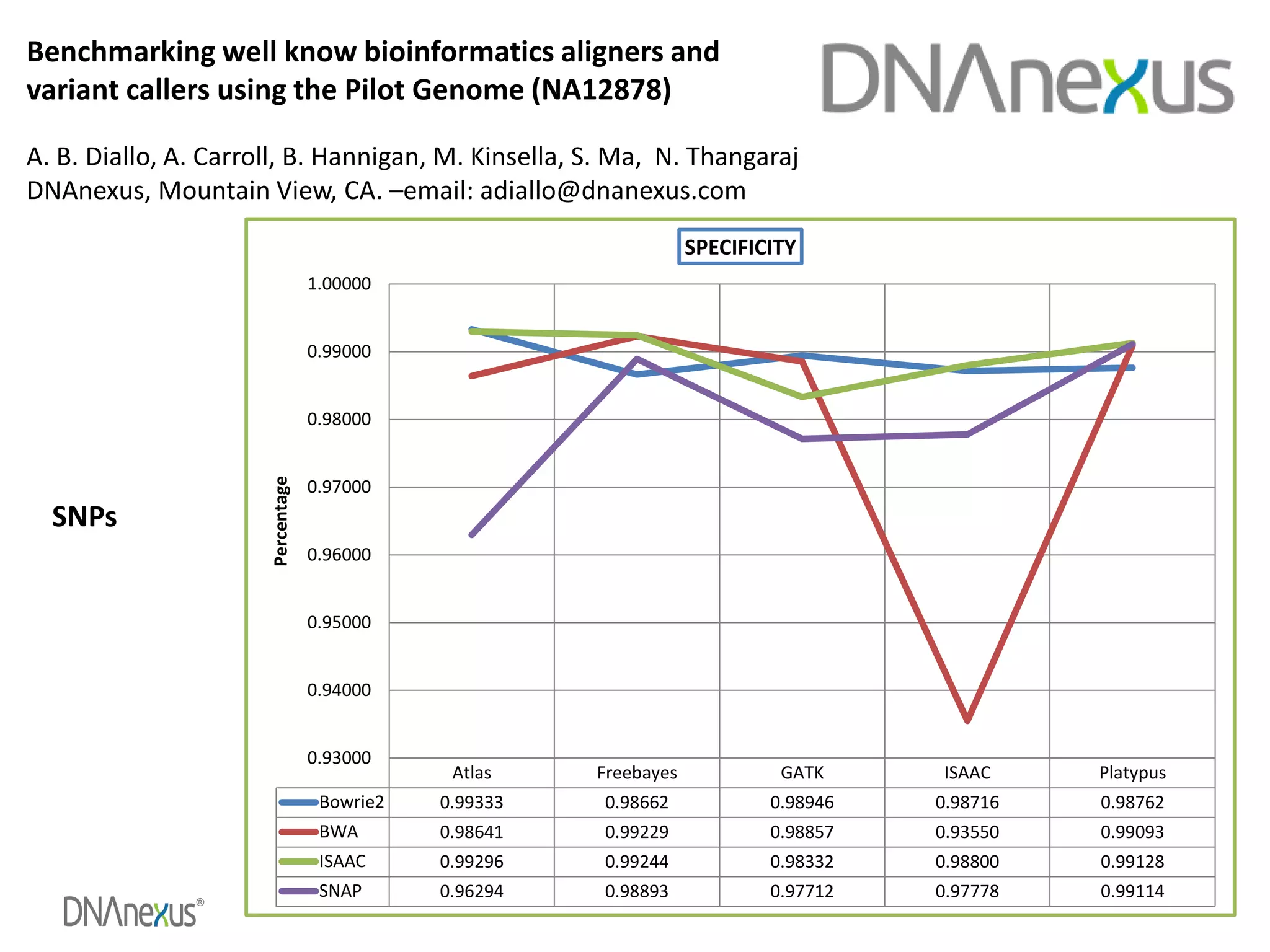
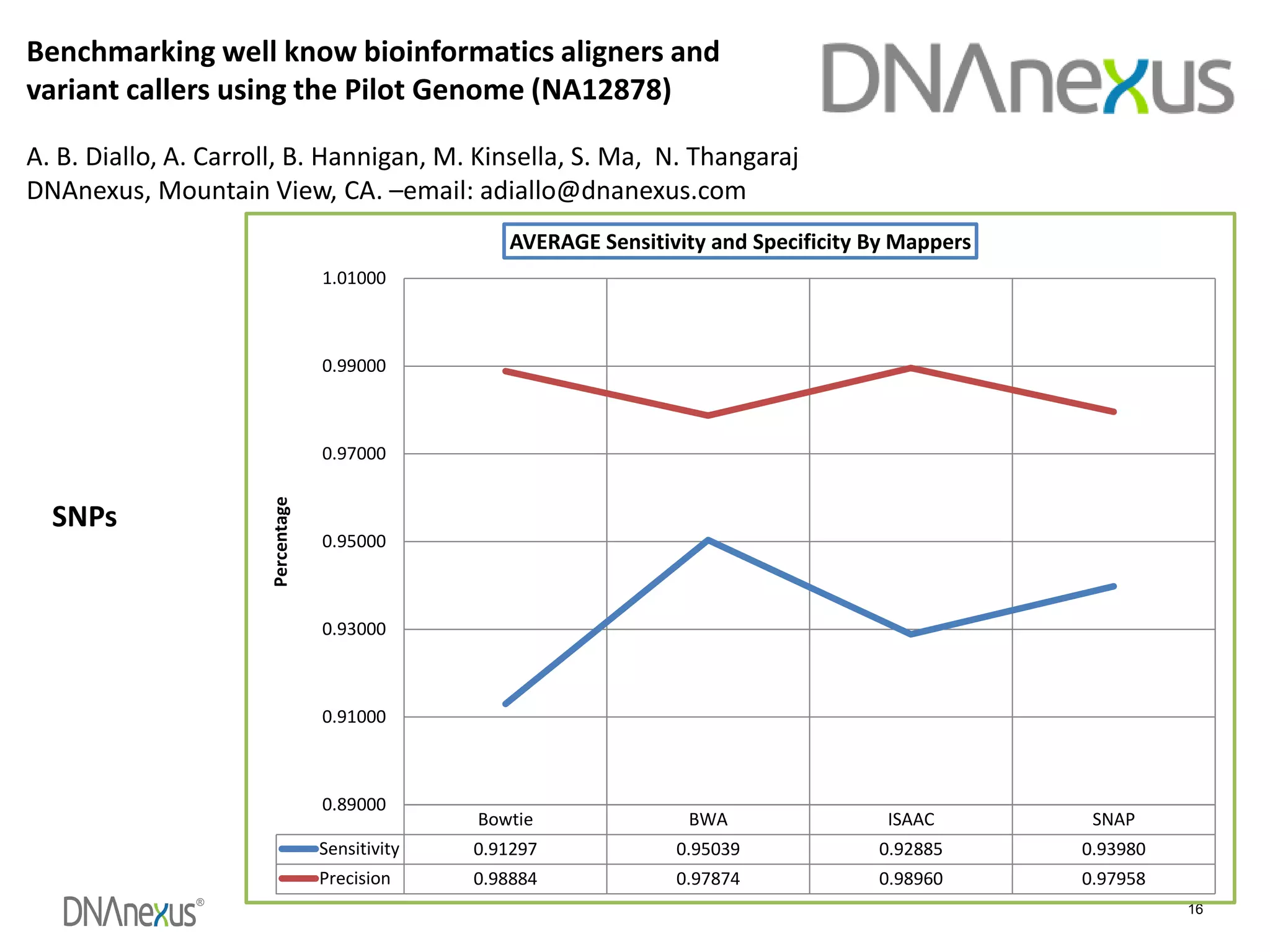
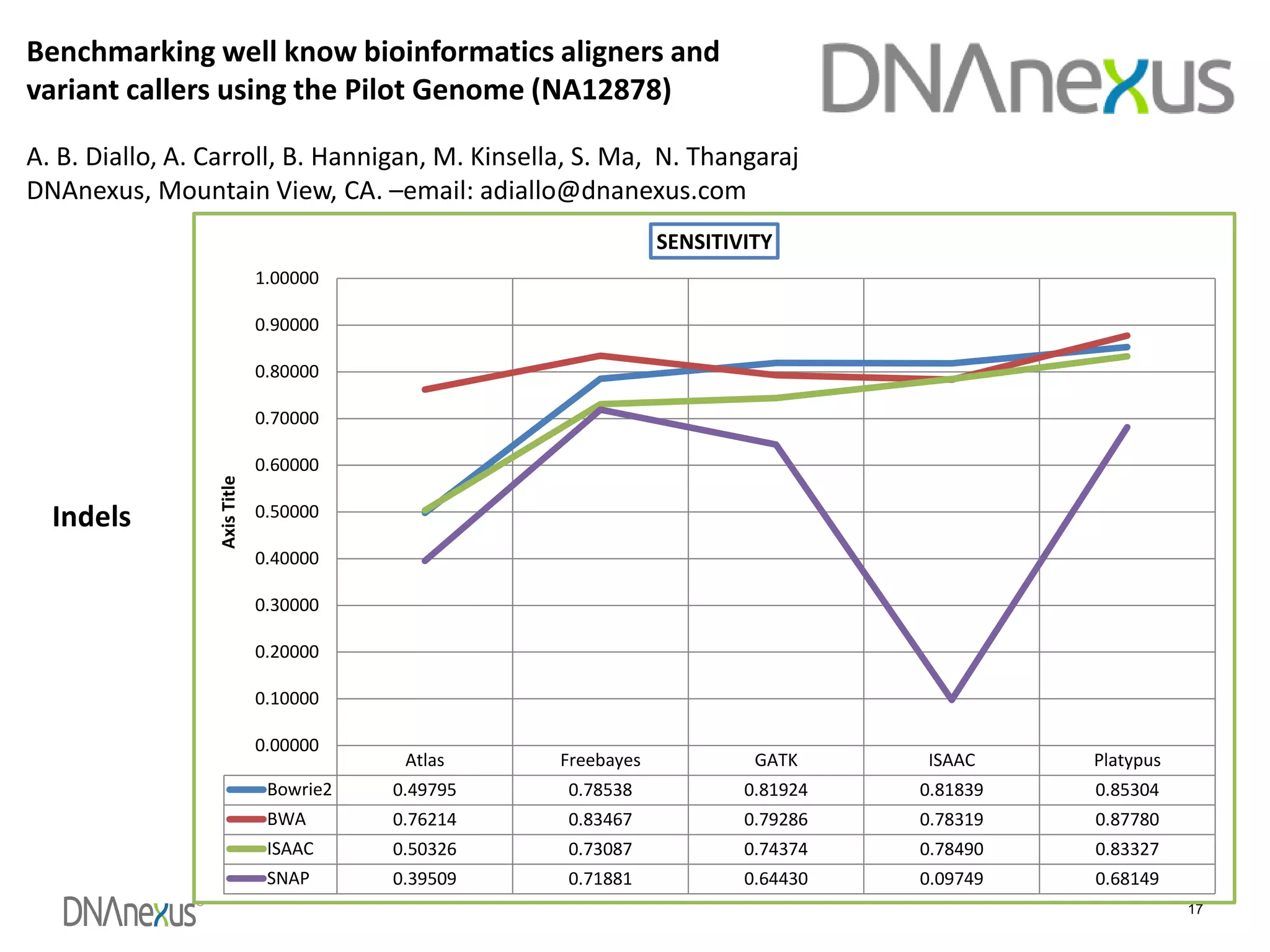
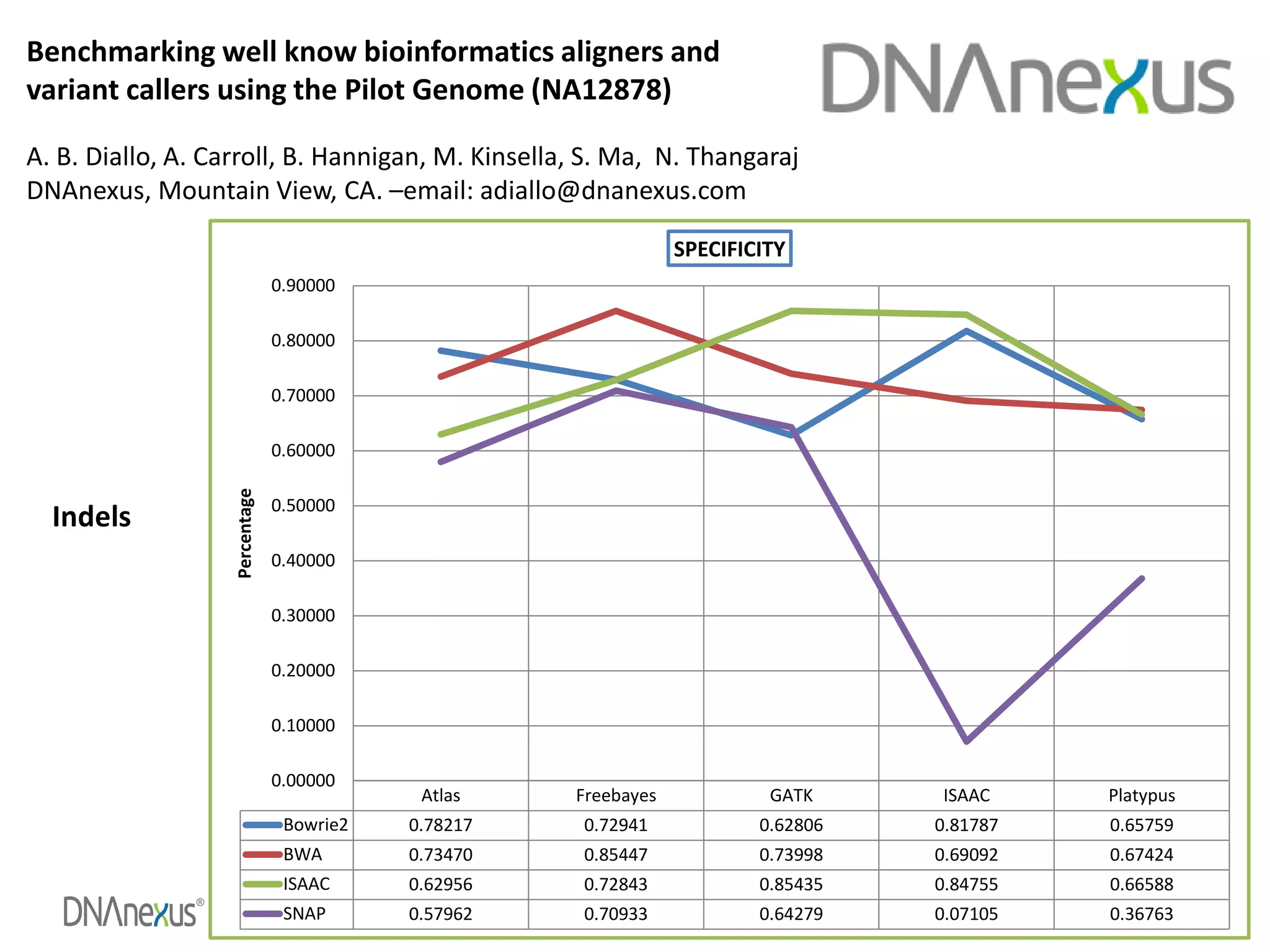
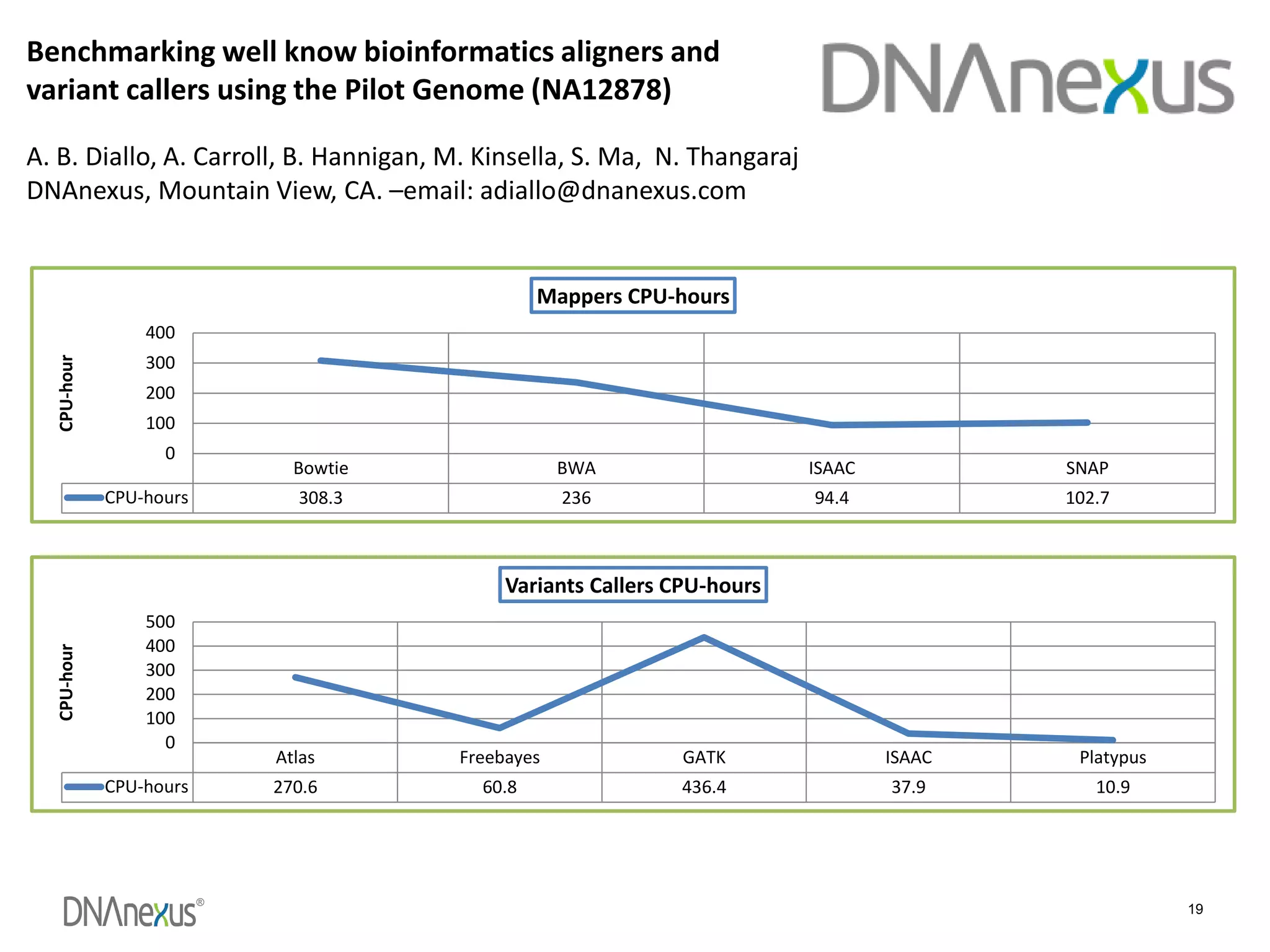
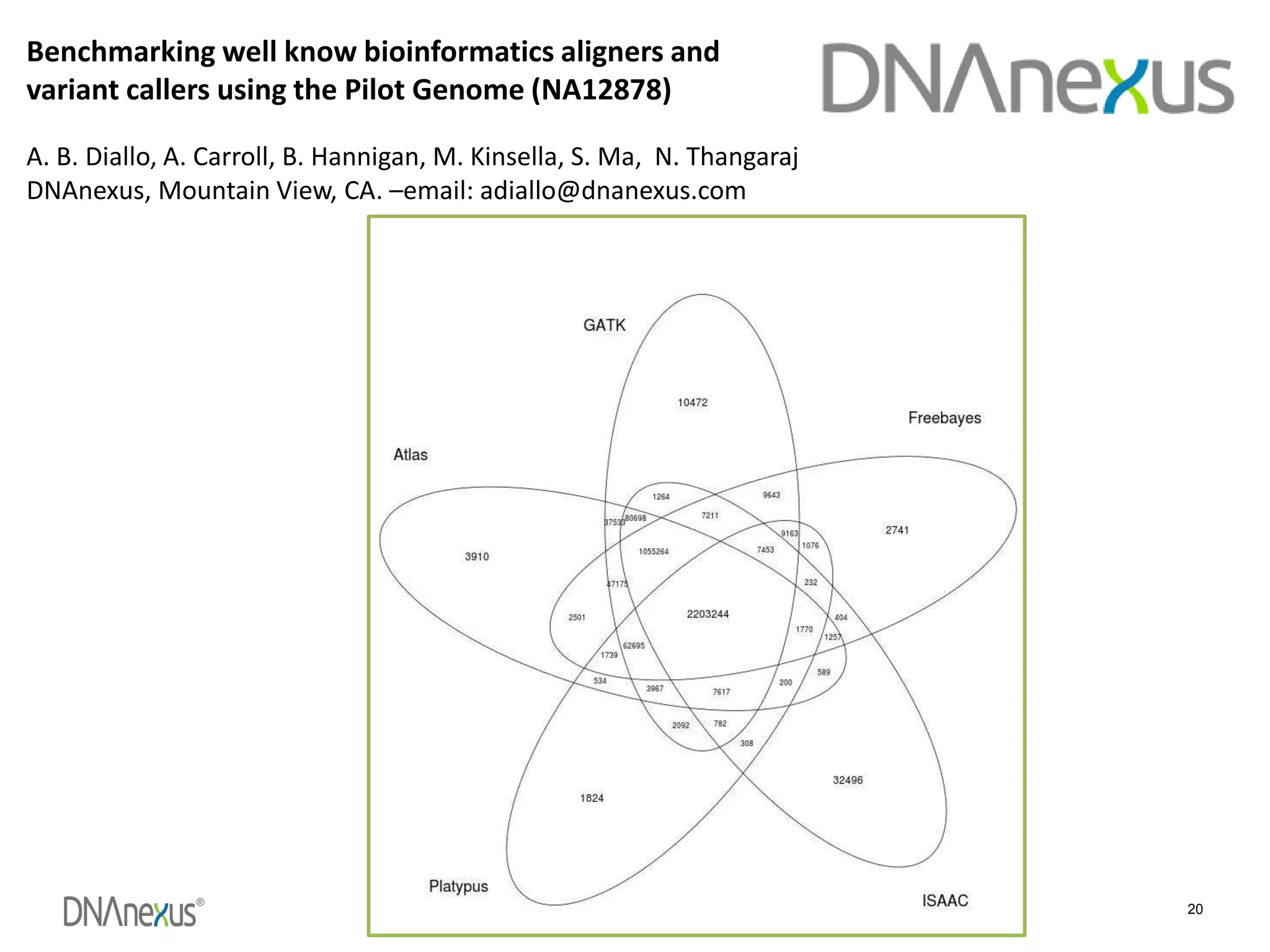
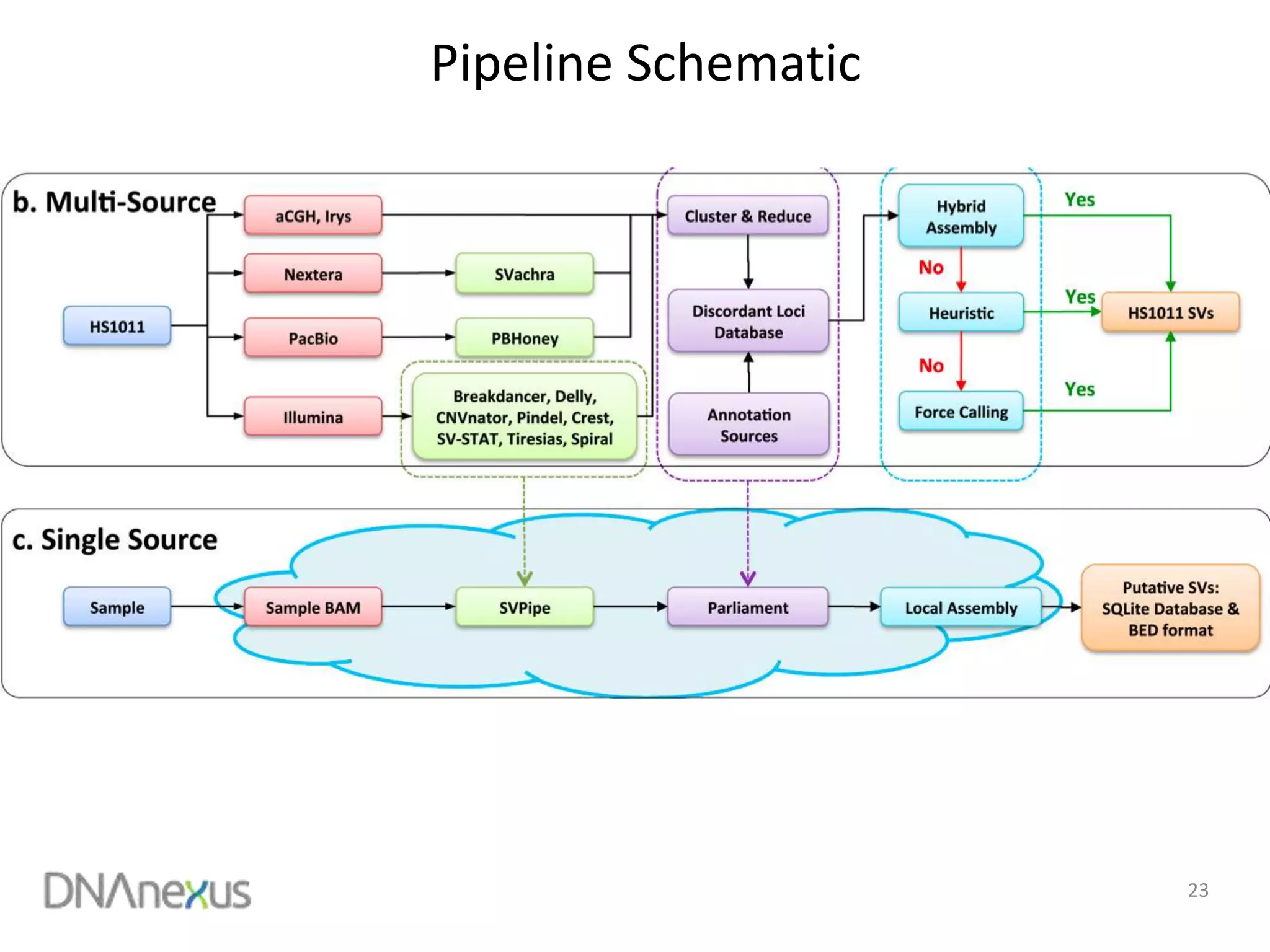
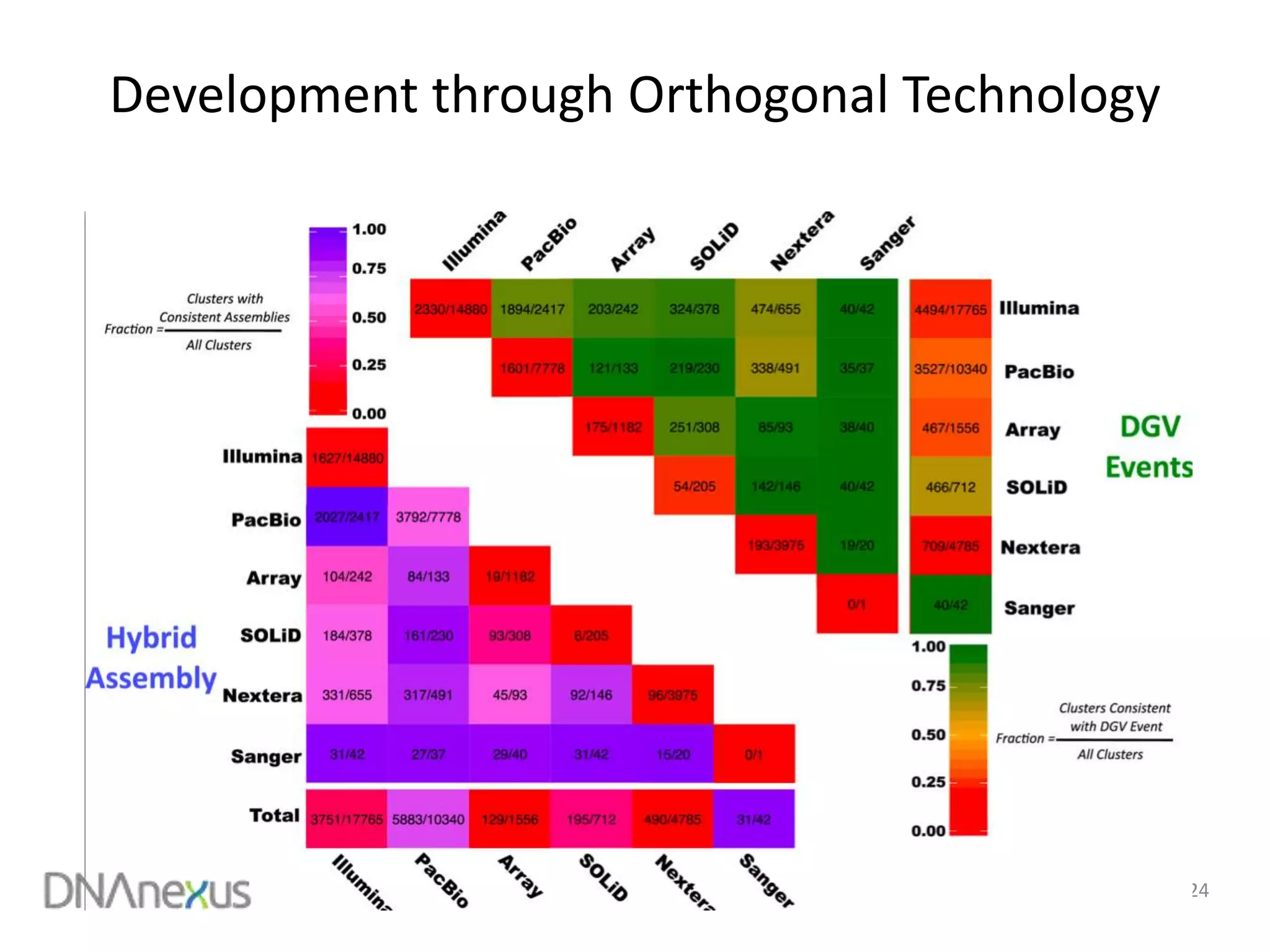
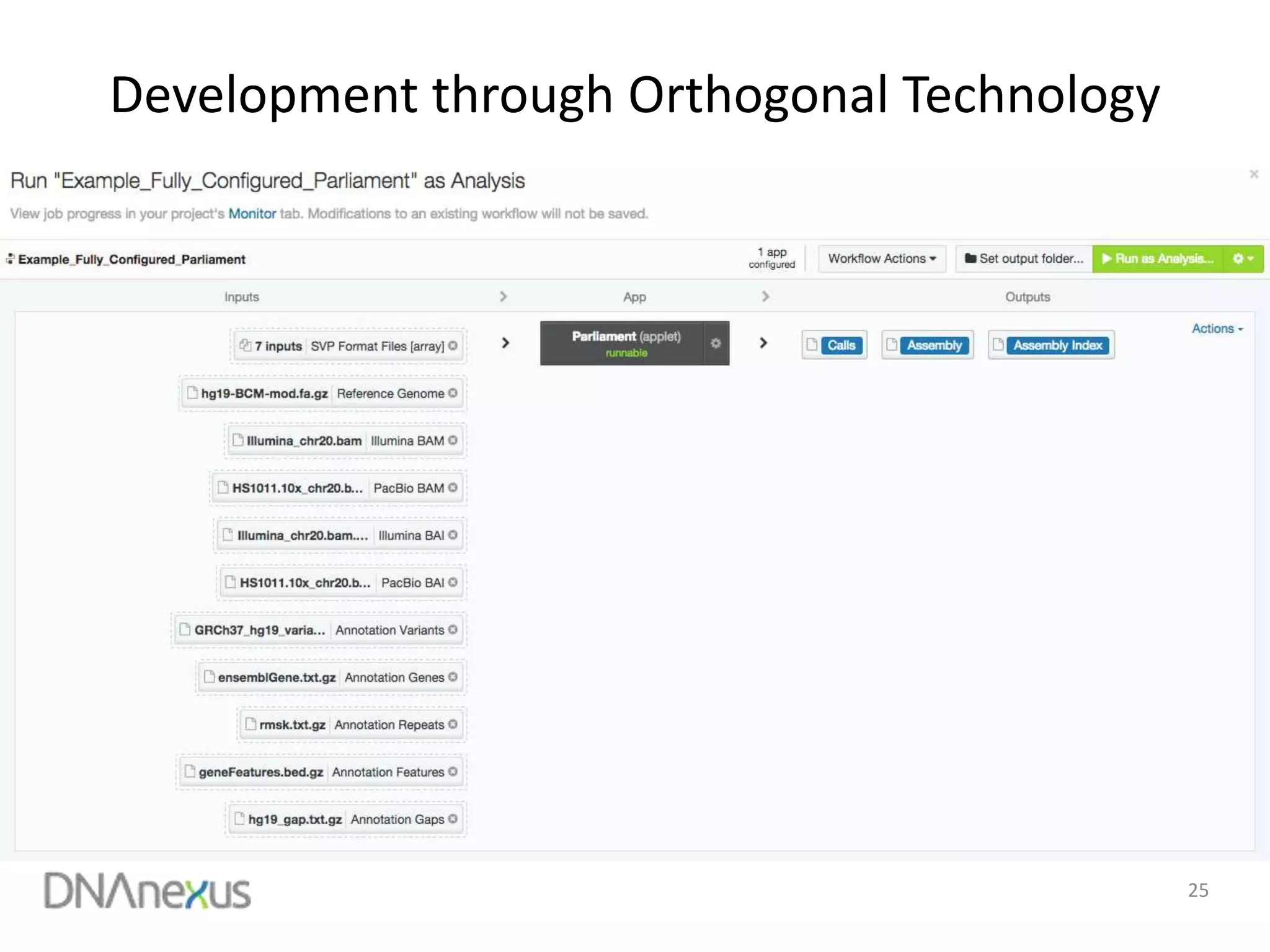
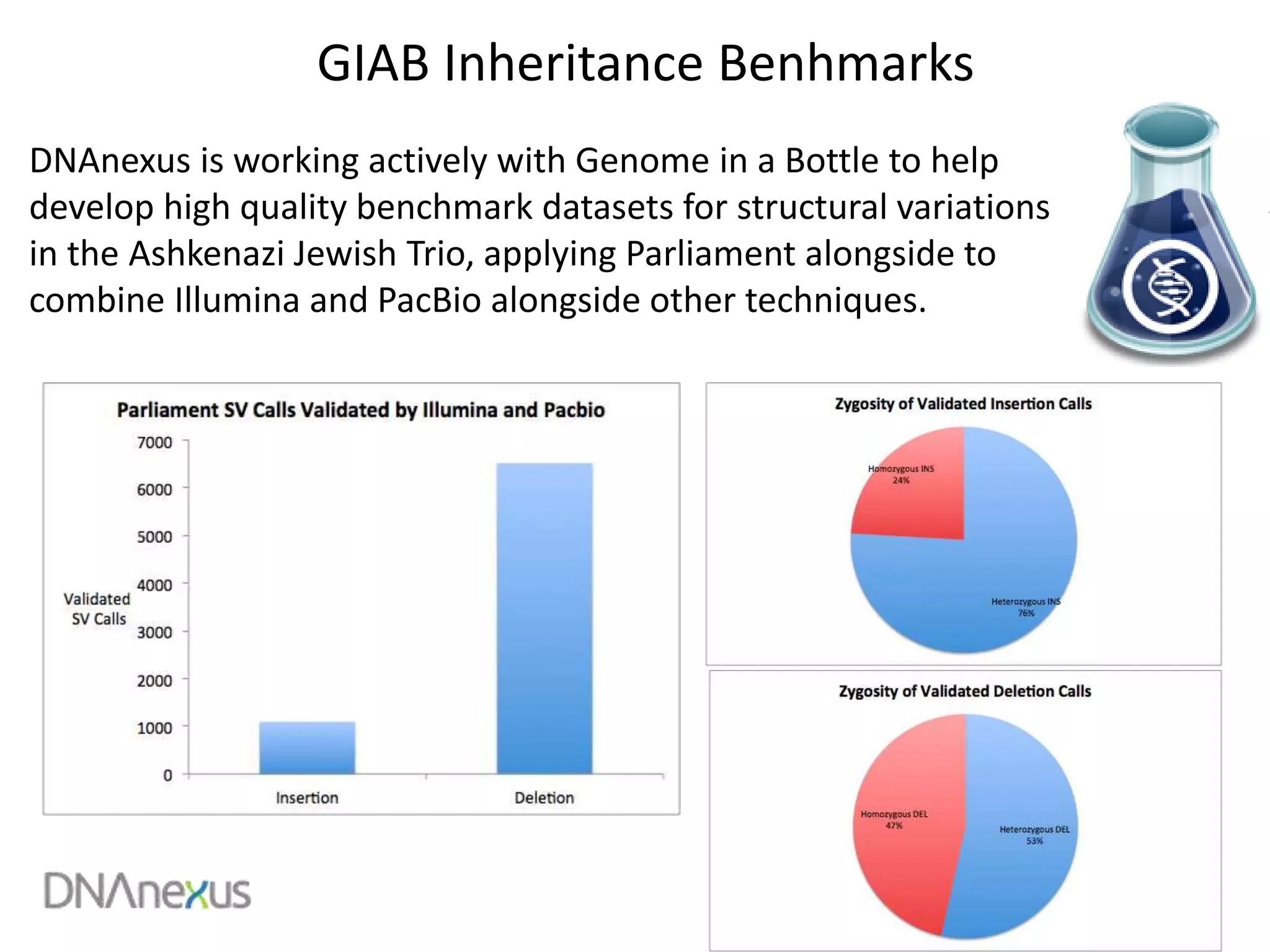
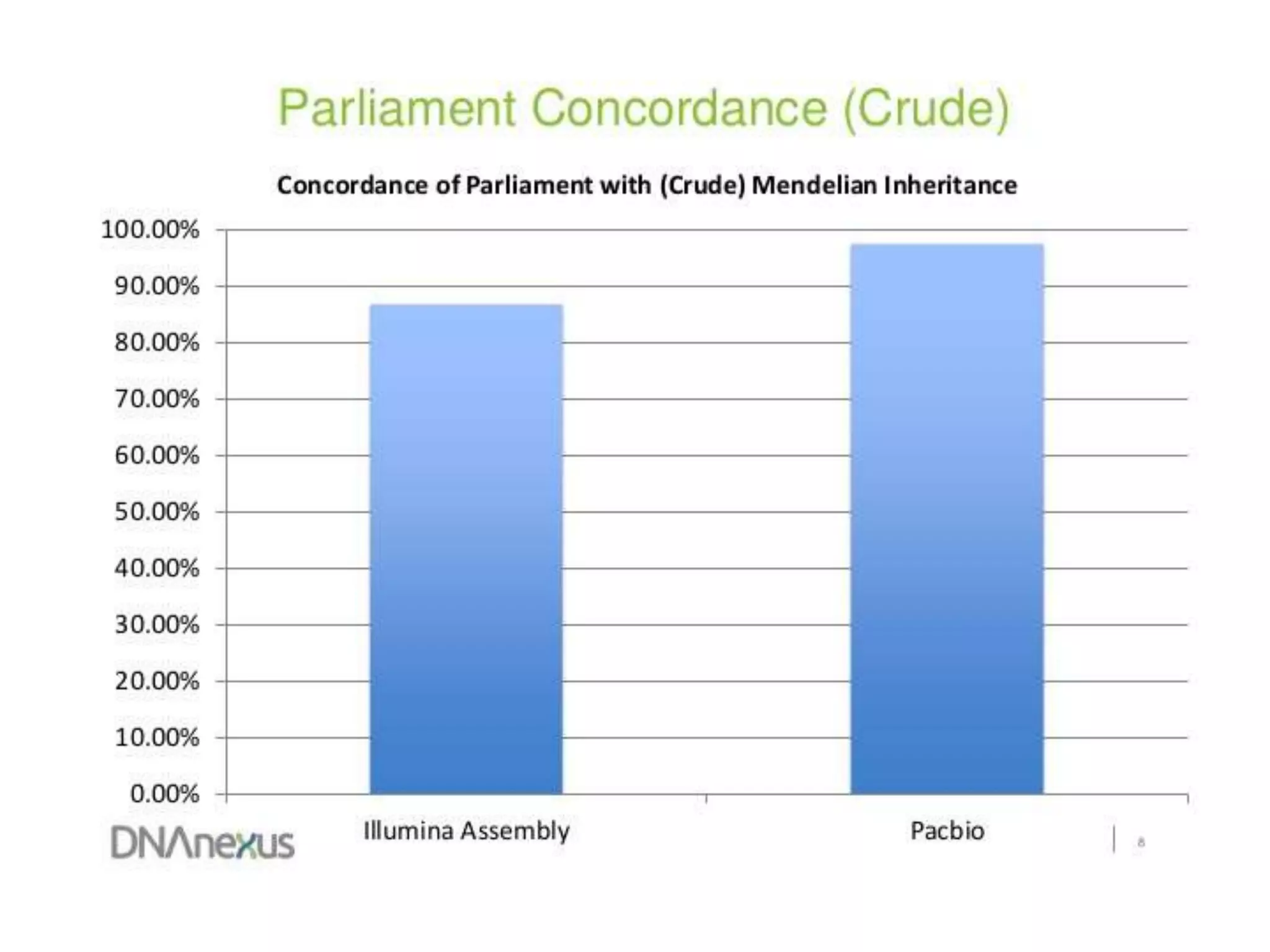
The document discusses using Genome in a Bottle (GIAB) data on DNAnexus cloud platform. It describes two examples: 1) Comparing different mapper and variant caller combinations using GIAB pilot genome data. Benchmarking shows BWA and GATK Haplotype Caller performed best. 2) Assessing structural variation detection in the Ashkenazi Jewish Trio, combining data from Illumina and PacBio sequencing. DNAnexus is working with GIAB to develop benchmark datasets for structural variants.