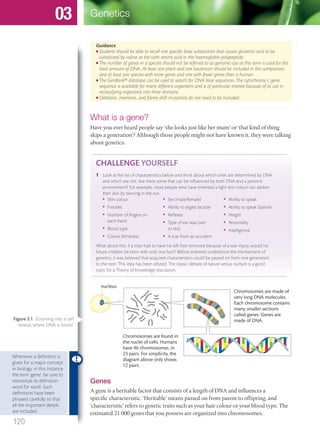
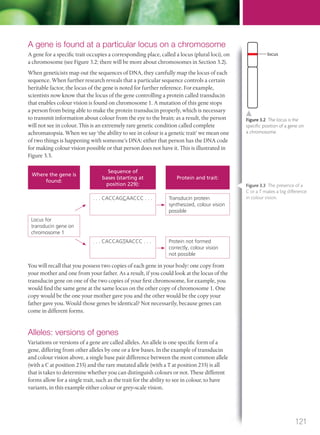
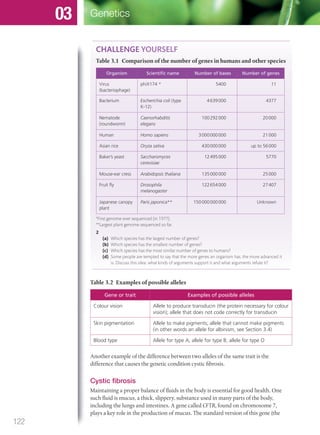
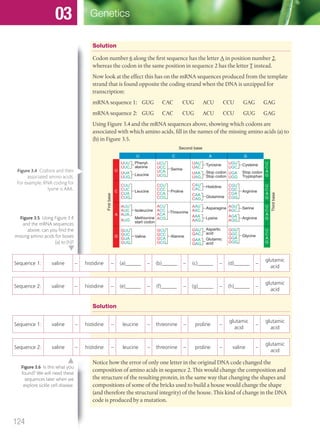
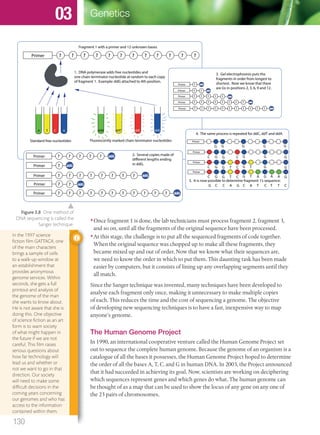
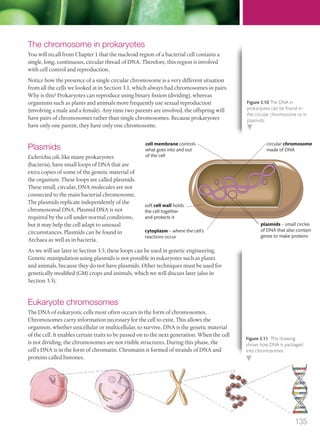
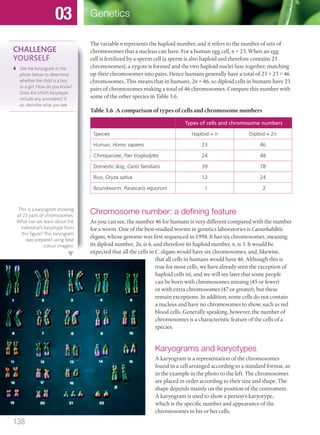
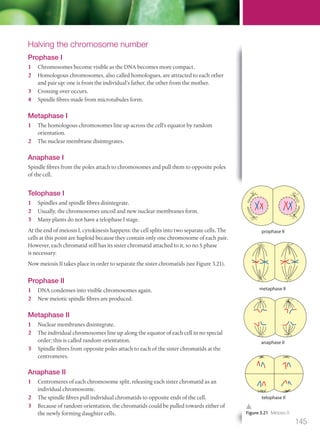
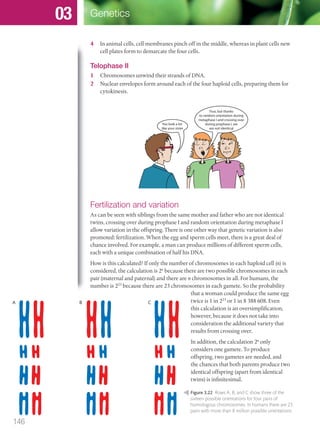
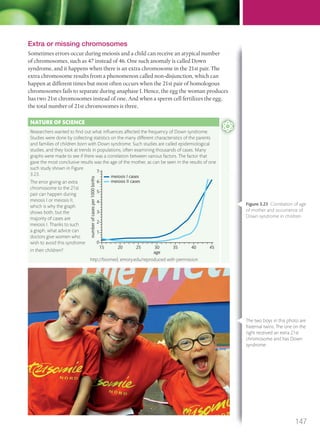
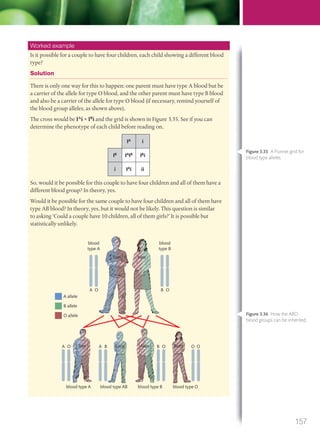
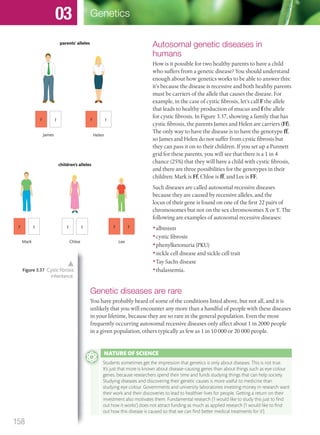
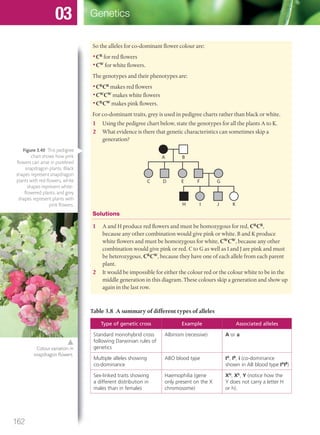
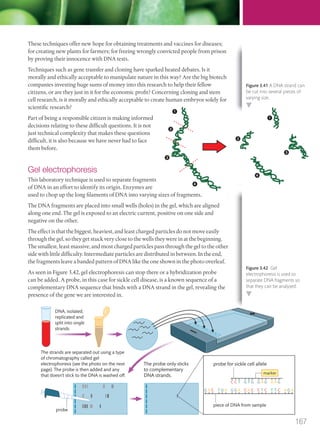
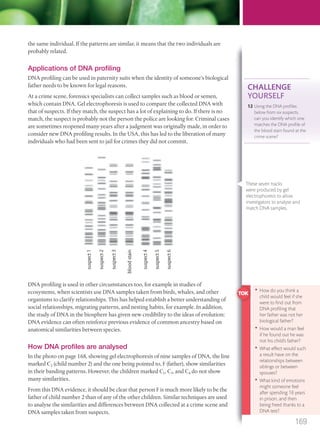
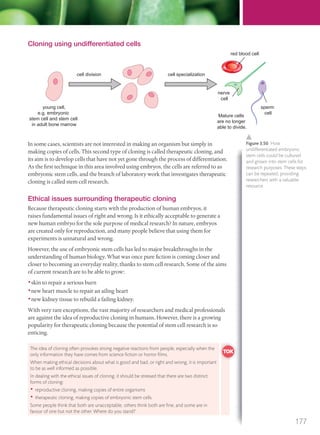
The document discusses genetics and genes. It states that chromosomes carry genes in a linear sequence shared by members of a species. Genes occupy specific positions on chromosomes and come in different alleles. Biologists have developed techniques to artificially manipulate DNA, cells, and organisms.