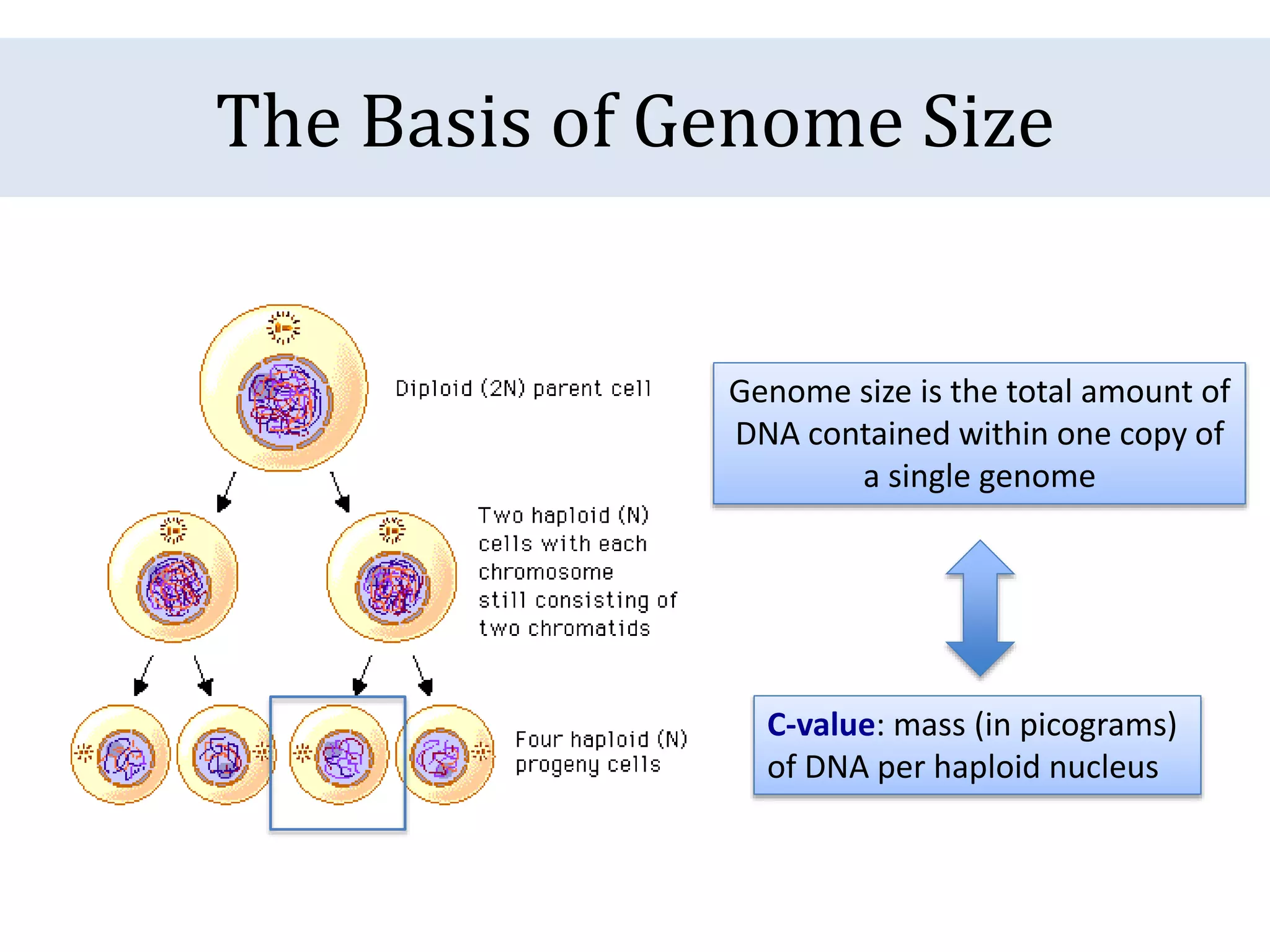
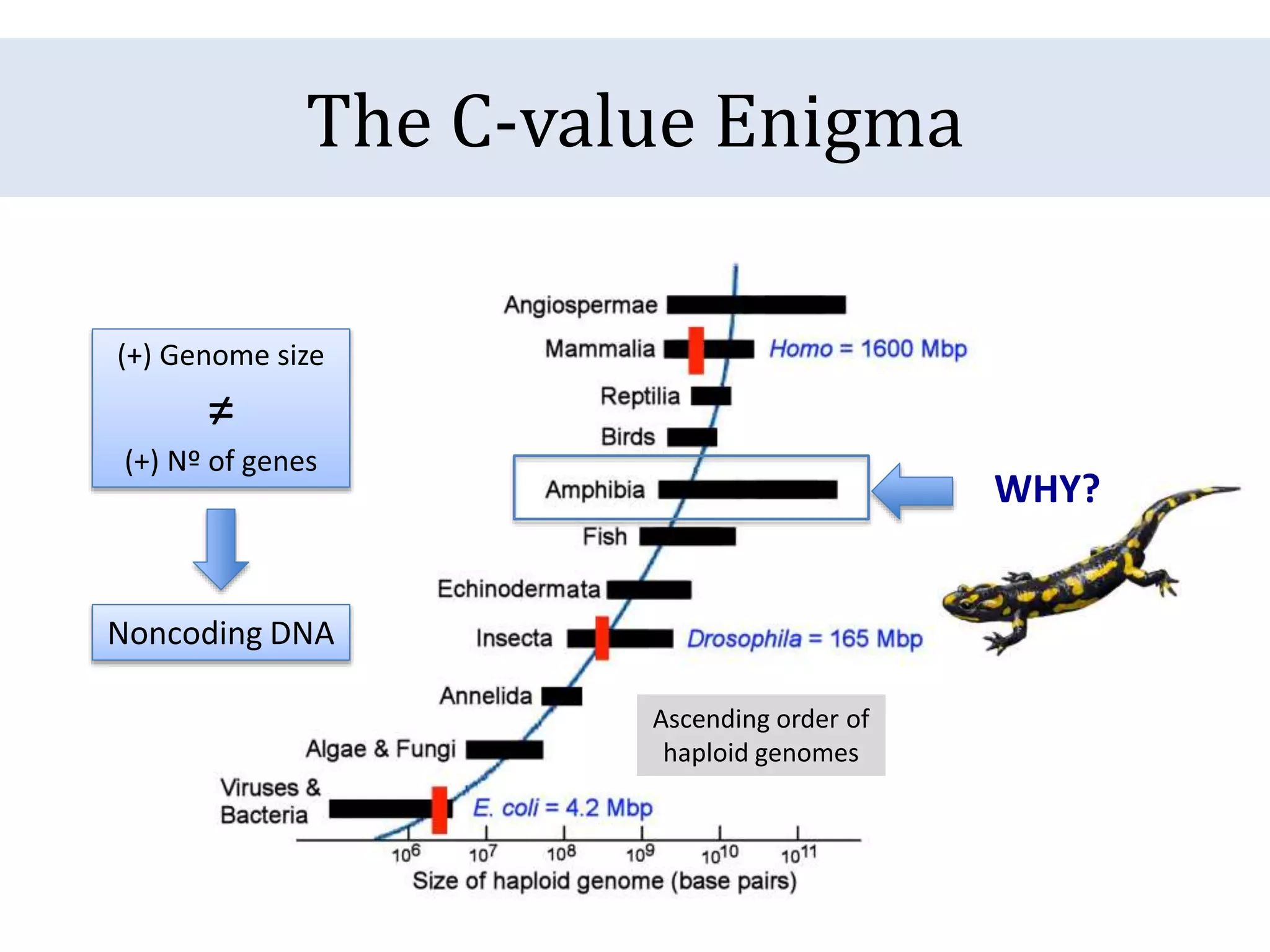
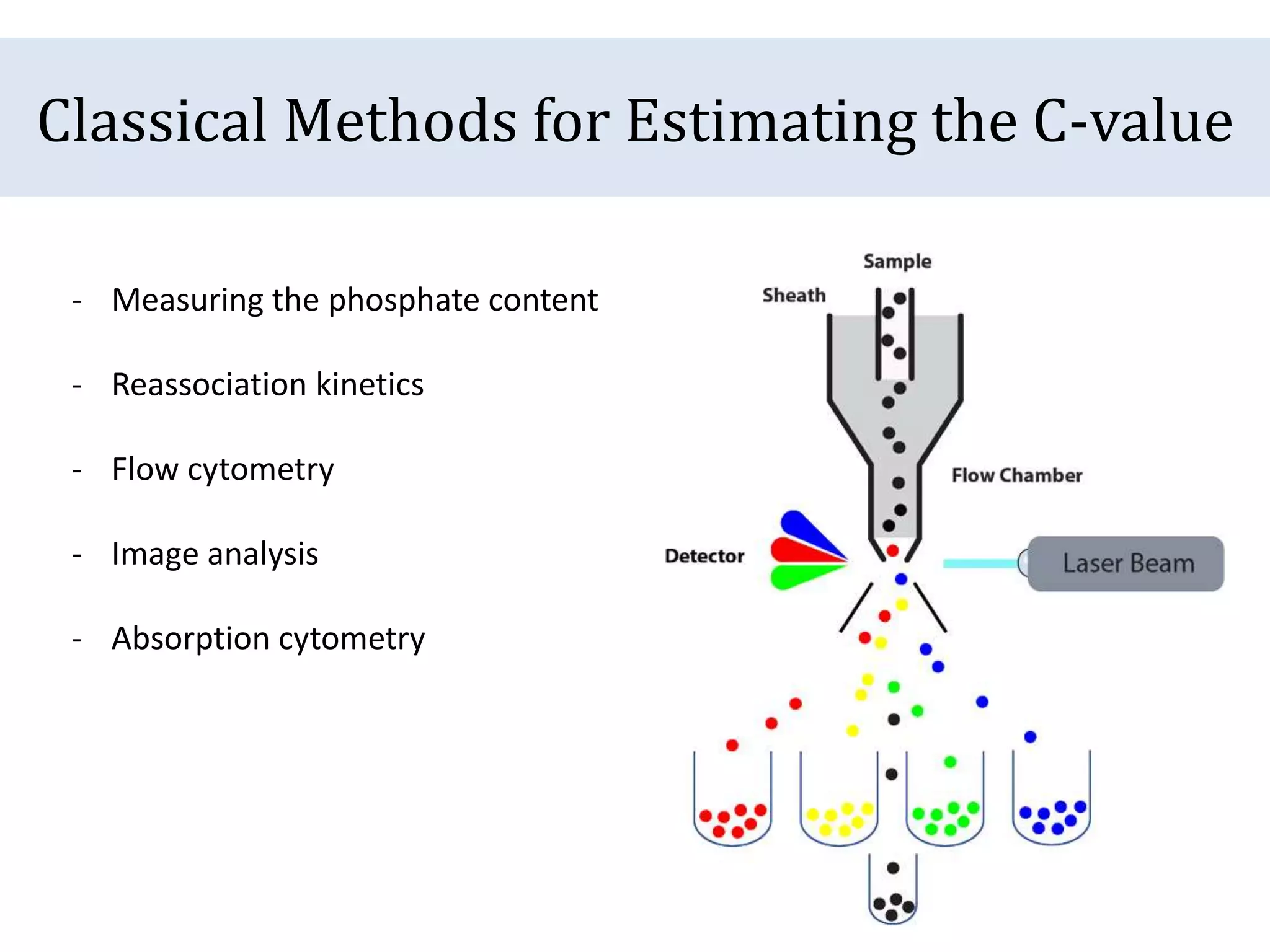
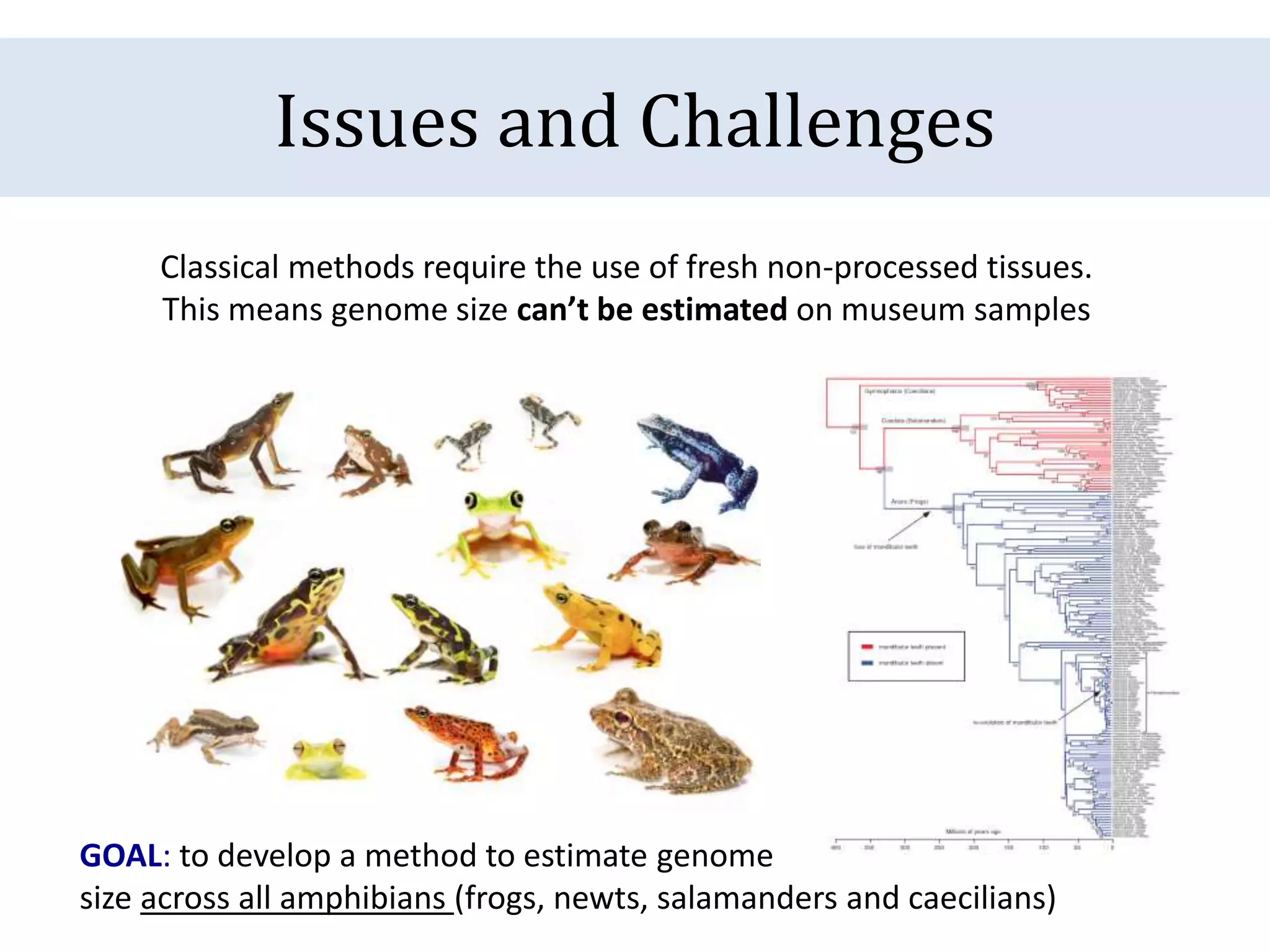
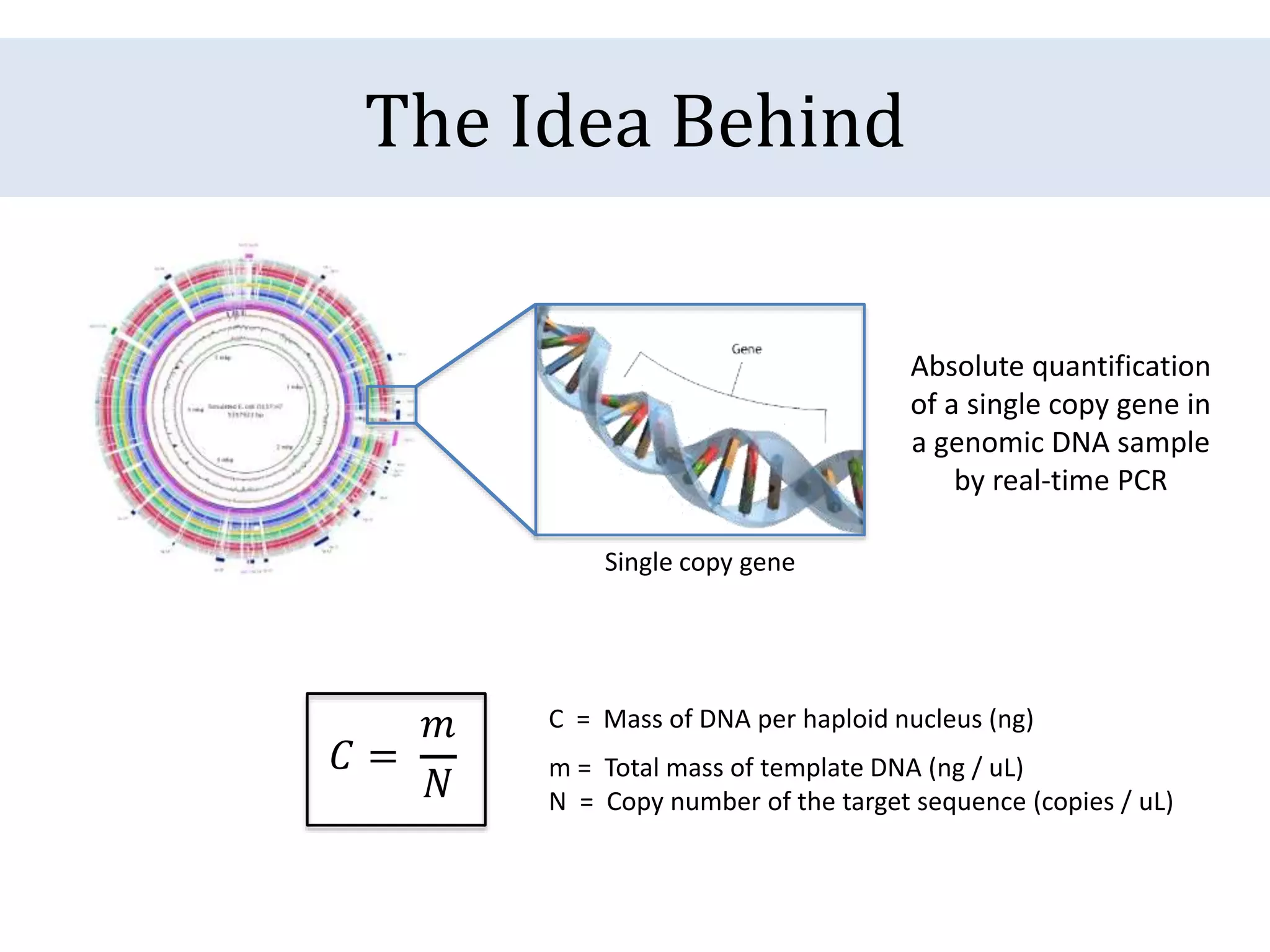
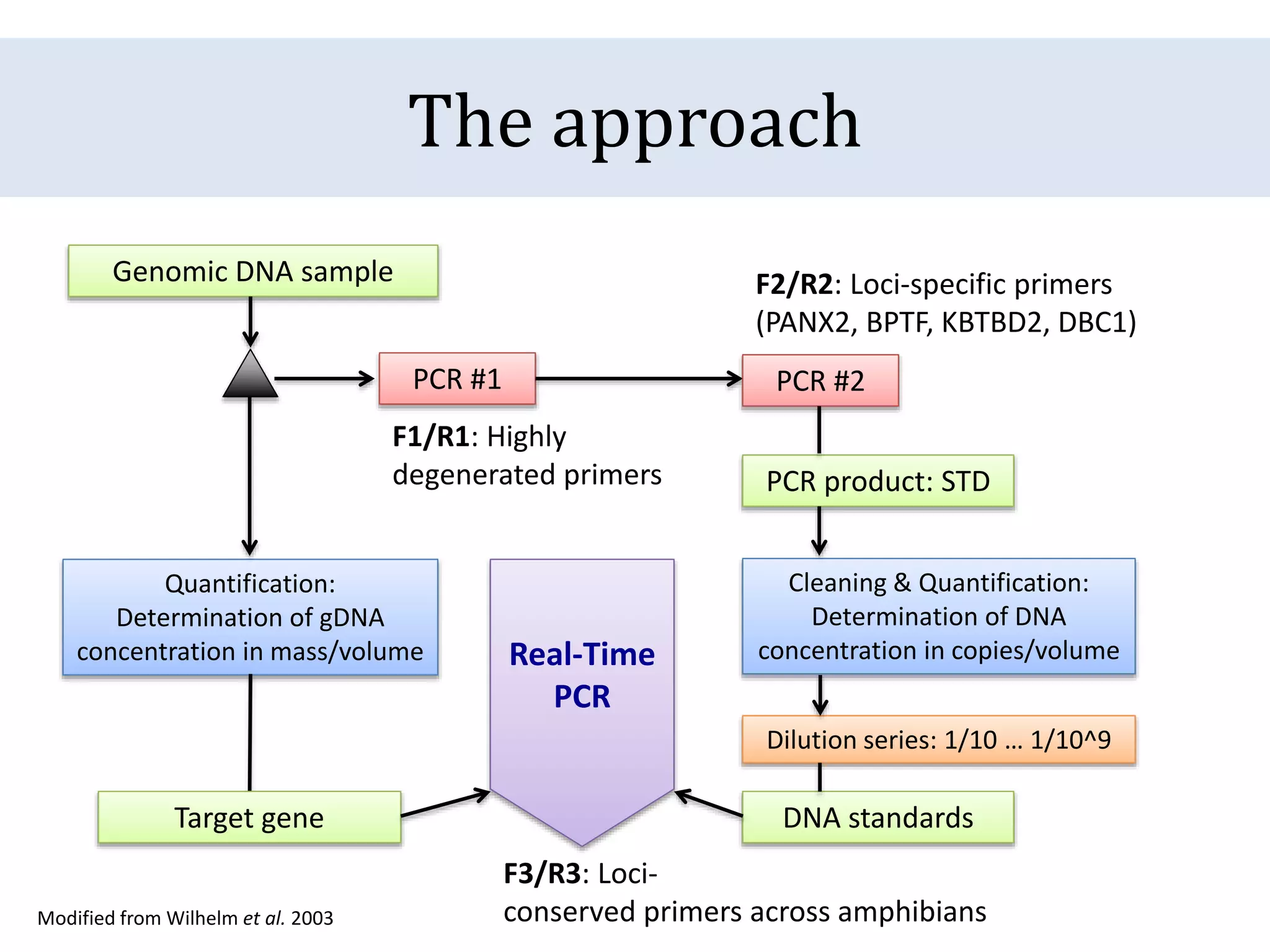
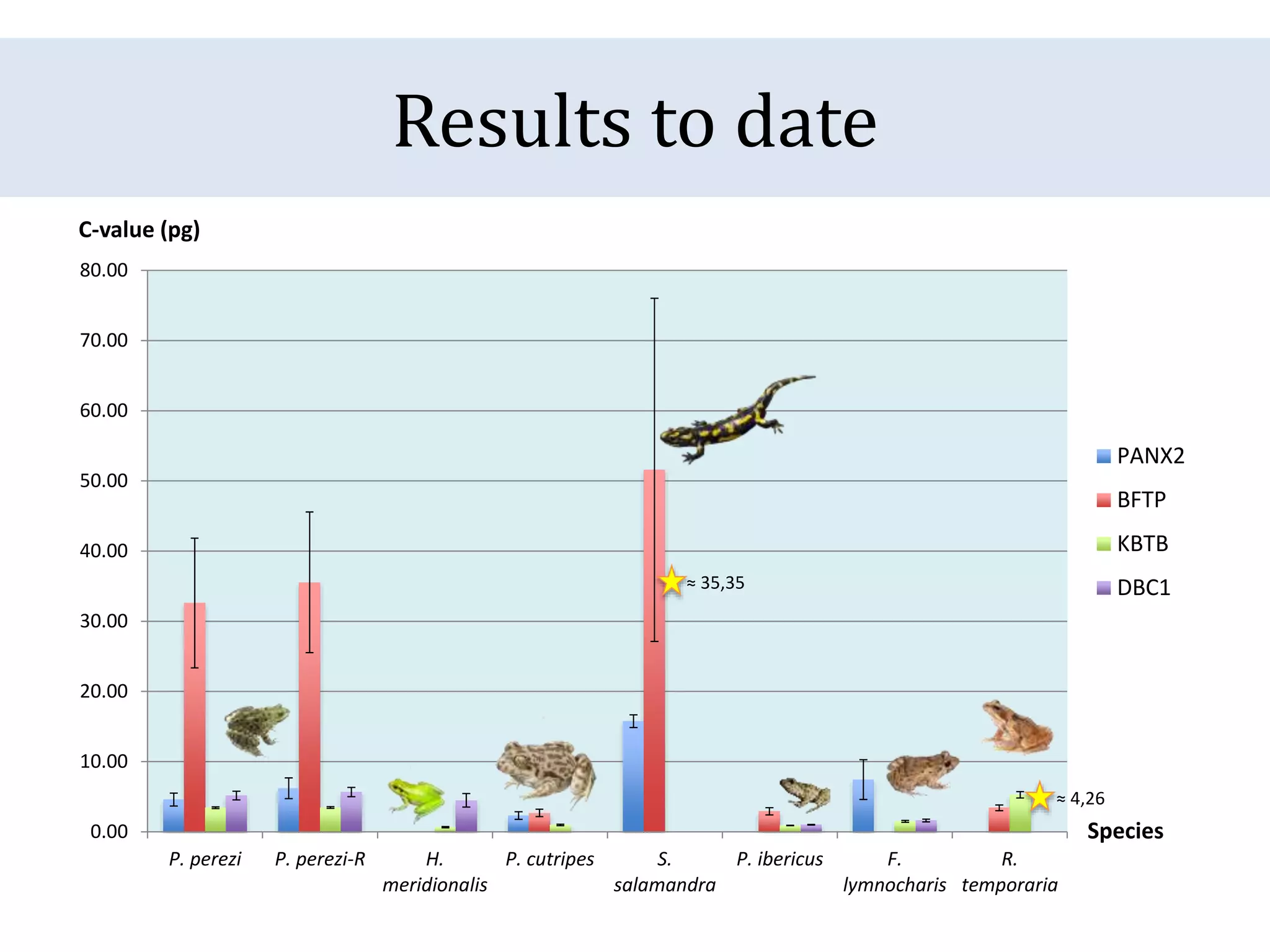
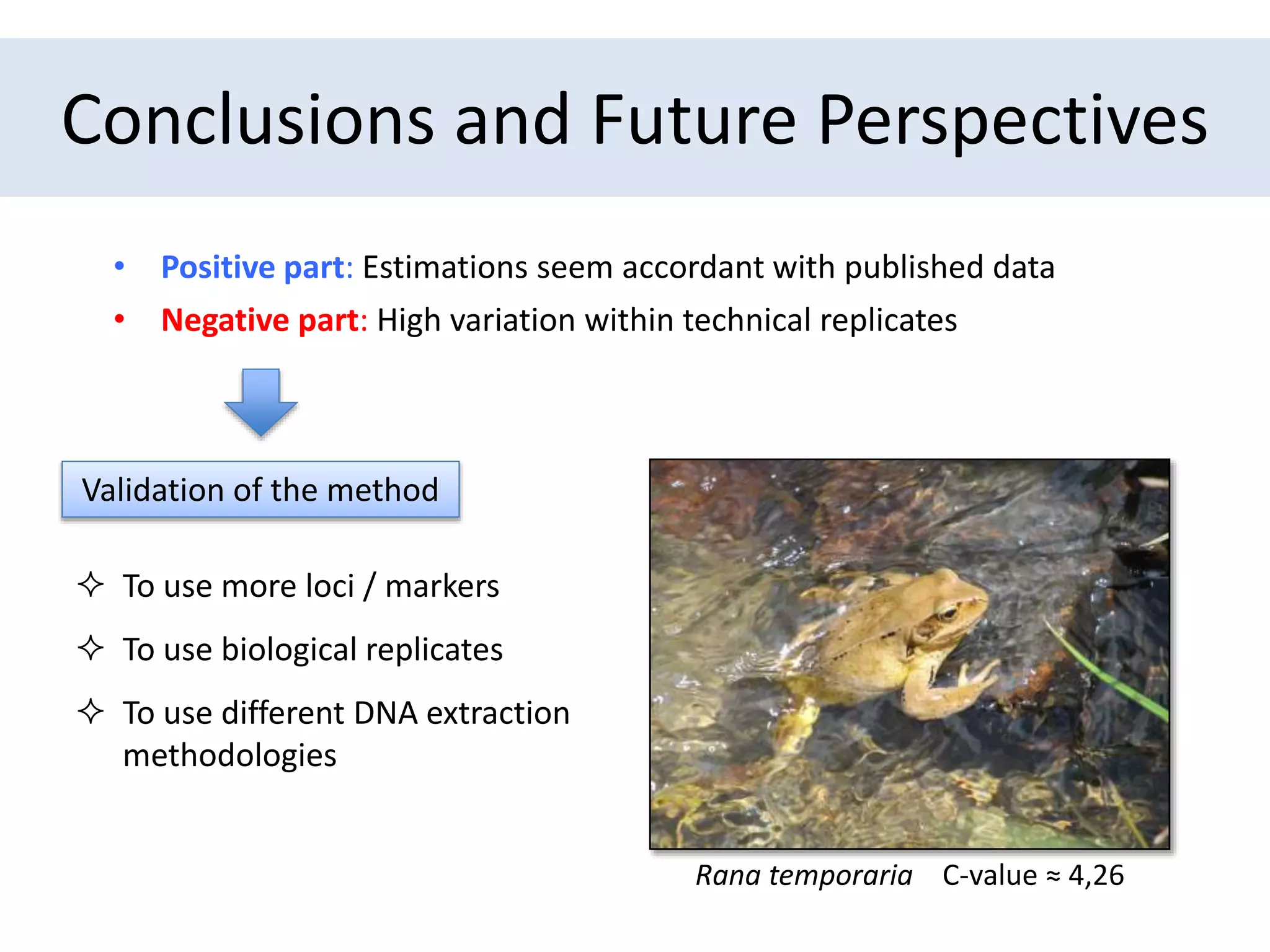
The document discusses a method for estimating genome size in amphibians using real-time PCR, emphasizing the importance of genome size in sequencing projects. It outlines classical challenges in measuring c-values and presents an innovative approach utilizing single copy genes for quantification. Preliminary results show alignment with existing data but highlight the need for further validation through additional loci and biological replicates.