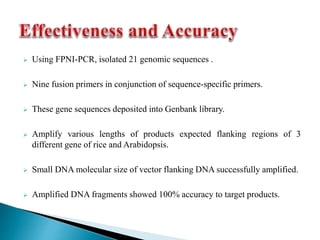
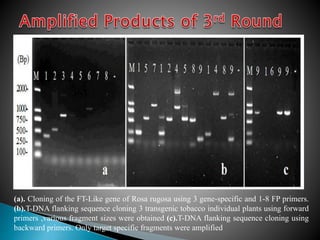
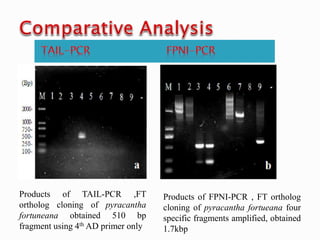
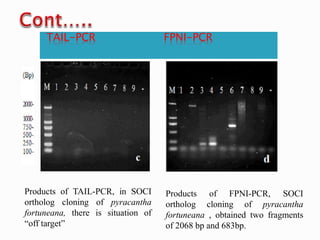
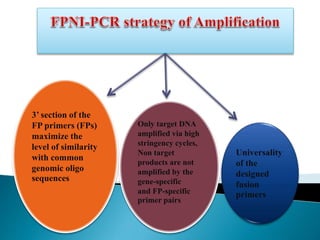
The document describes a method called Fusion Primer-based Nested Insertion PCR (FPNI-PCR) for amplifying unknown genomic DNA sequences. FPNI-PCR uses gene-specific primers and fusion primers containing an arbitrary sequence and known adaptor region. It was able to isolate 21 genomic sequences from 7 plant species in under 4 hours with 2 rounds of PCR and yielded fragments over 1kb in size. FPNI-PCR provided 100% specific amplification of target products and did not require DNA dilution steps, making it more efficient than other methods like TAIL-PCR.


![ Extraction of Genomic DNA of seven “Rosaceace” species.
Genomic DNA of tobacco, Petunia hybrida, Arabidopsis and
rice. [20]
From young leaves by NaOH fast extraction. [21]](https://image.slidesharecdn.com/final1-150629162759-lva1-app6892/85/Nested-PCR-Fusion-primers-Integrated-PCR-4-320.jpg)


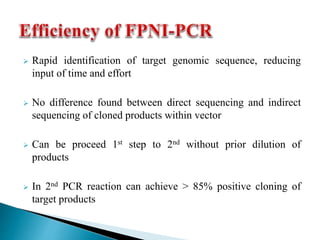

![ Yield small DNA fragments
2 rounds of laborious DNA dilutions
Uneconomical for primer designing
Just 60% of reactions yielded specific
products [3]
Take a whole working day
Demand only extremely modest
levels of quantity and purity of the
template DNA
FPNI-PCR reactions yielded DNA
fragments larger than 1.0 kb [6]
Less complicated and easy to
manipulate
100% specific product amplification
through high and low annealing T
[1,3]
42-45 cycles take only 2 hours
Demand only extremely modest
levels of quantity and purity of the
template DNA](https://image.slidesharecdn.com/final1-150629162759-lva1-app6892/85/Nested-PCR-Fusion-primers-Integrated-PCR-17-320.jpg)

![1. [1] Liu YG, Mitsukawa N, Oosumi T, Whittier RF: Efficient isolation and mapping of
Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The
Plant Journal 1995, 8:457-463.
2. [2] Ji JB, Braam J: Restriction Site Extension PCR: A Novel Method for HighThroughput
Characterization of Tagged DNA Fragments and Genome Walking. Plos One 2010, 5:1-5.
3. [3] Liu YG, Chen YL: High-efficiency thermal asymmetric interlaced PCR for
amplification of unknown flanking sequences. BioTechniques 2007, 43:649-656.
4. [4] Reddy PS, Mahanty S, Kaul T, Nair S, Sopory SK, Reddy MK: A highthroughput
genome-walking method and its use for cloning unknown flanking sequences. Analytical
Biochemistry 2008, 381:248-253.
5. [15] Liu FY, Ma B, Zhao Y: Characterization of the gene encoding glycoprotein C of duck
enteritis virus. Virus Genes 2008, 37:328-332.
6. [20] Yang C, Zhang J, Xu Q, Xiong C, Bao M: Establishment of AFLP technique and
assessment of primer combinations for Mei Flower. Plant Molecular Biology Reporter
2005, 23:790-791.
7. [21] He YH, Ning GG, Sun YL, Qi YC, Bao MZ: Identification of a SCAR marker linked
to a recessive male sterile gene (Tems) and its application in breeding of marigold (Tagetes
erecta L.). Plant breeding 2009, 128:92-96](https://image.slidesharecdn.com/final1-150629162759-lva1-app6892/85/Nested-PCR-Fusion-primers-Integrated-PCR-19-320.jpg)
![1. [20] Yang C, Zhang J, Xu Q, Xiong C, Bao M: Establishment of AFLP
technique and assessment of primer combinations for Mei Flower. Plant
Molecular Biology Reporter 2005, 23:790-791.
2. [21] He YH, Ning GG, Sun YL, Qi YC, Bao MZ: Identification of a
SCAR marker linked to a recessive male sterile gene (Tems) and its
application in breeding of marigold (Tagetes erecta L.). Plant breeding
2009, 128:92-96](https://image.slidesharecdn.com/final1-150629162759-lva1-app6892/85/Nested-PCR-Fusion-primers-Integrated-PCR-20-320.jpg)