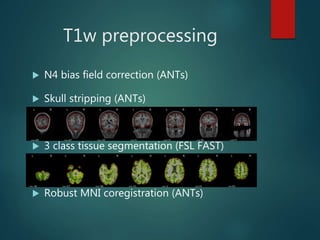
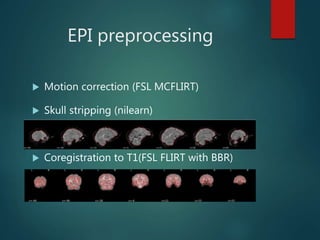
FMRIPREP is an fMRI preprocessing tool that provides robust, easy-to-use preprocessing of MRI data including denoising and normalization. It performs T1-weighted preprocessing like N4 bias field correction and tissue segmentation. It also performs EPI preprocessing including motion correction, skull stripping, coregistration to T1, and estimation of noise confounds. FMRIPREP outputs preprocessed data in native and MNI space. It is designed to be transparent and works on any BIDS-formatted dataset through either Docker or Singularity containers.