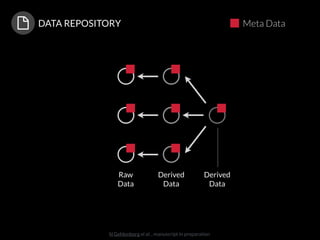
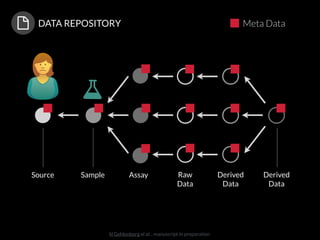
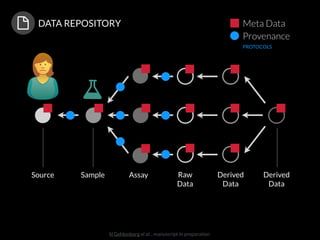
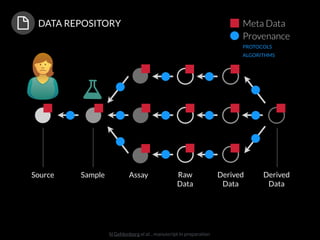
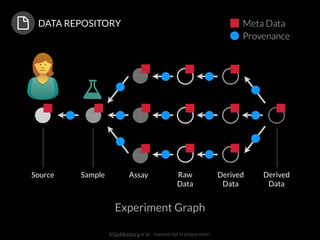
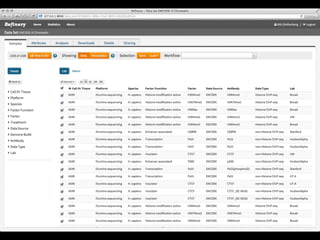
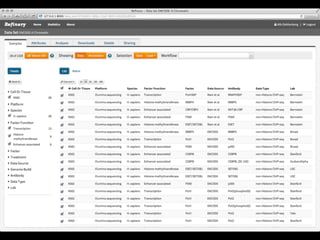
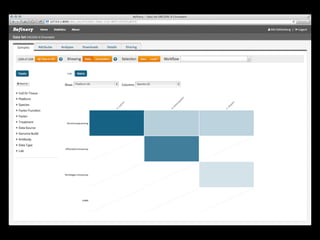
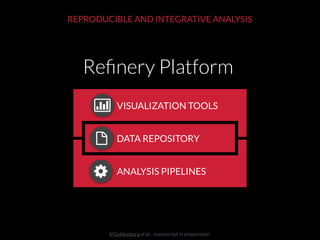
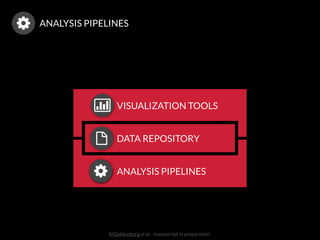
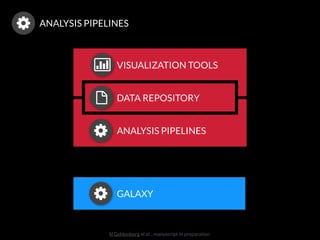
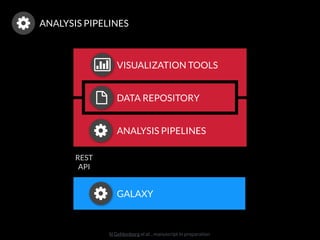
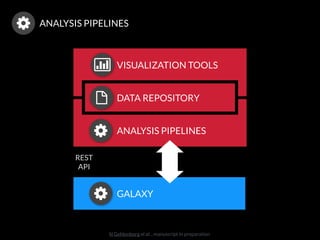
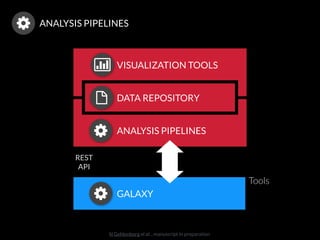
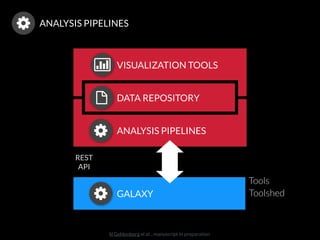
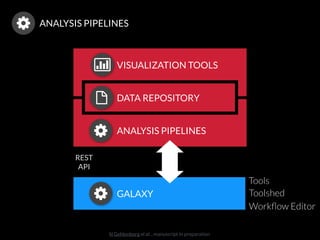
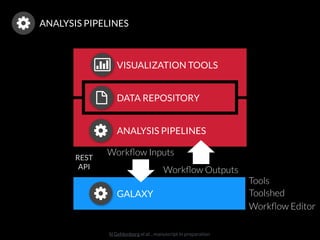
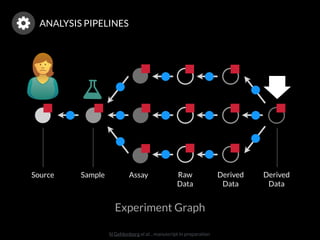
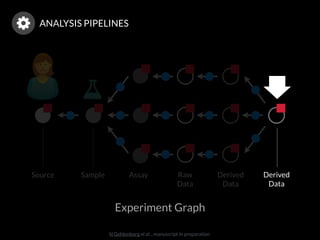
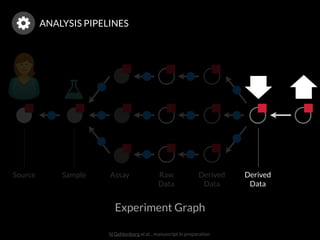
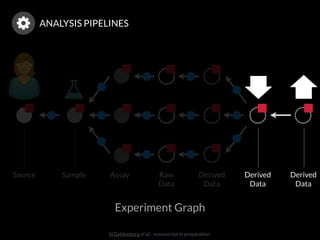
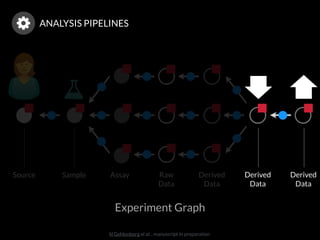
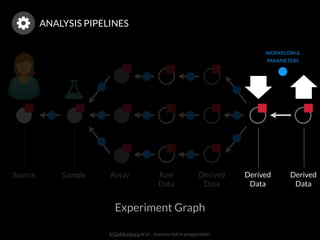
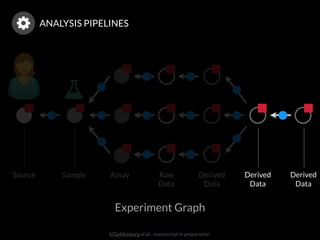
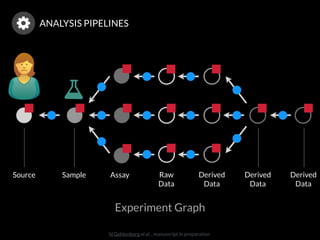
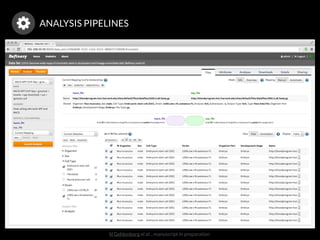
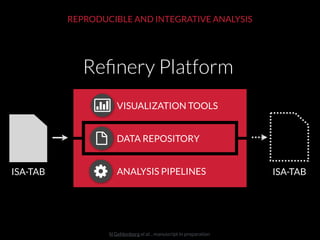
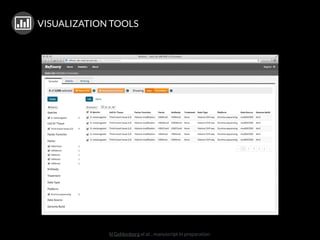
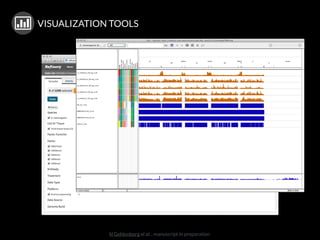
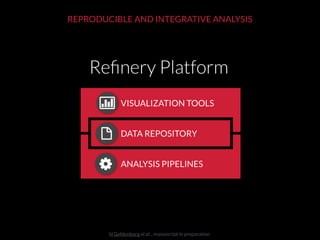
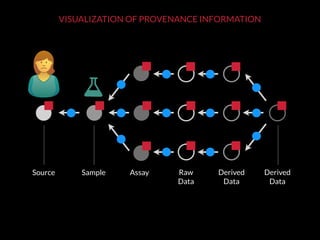
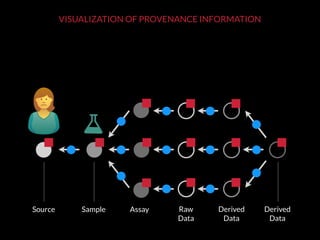
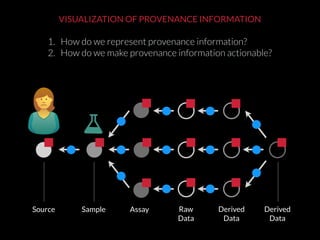
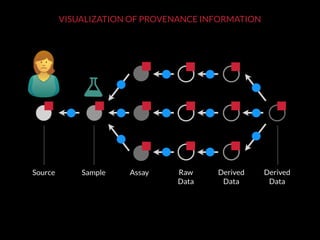
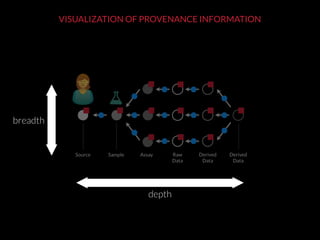
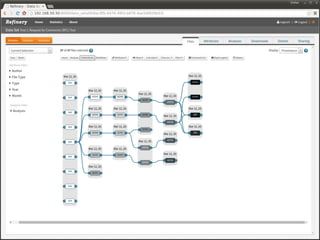
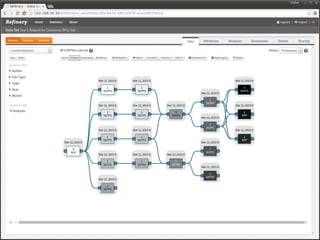
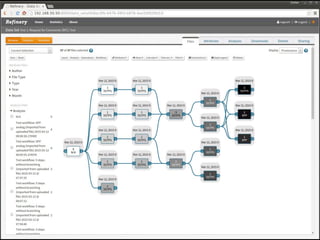
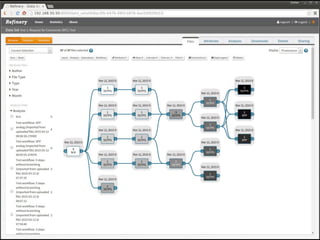
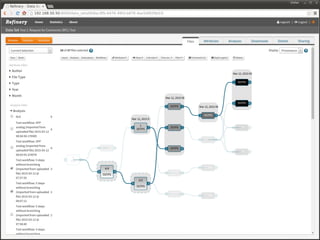
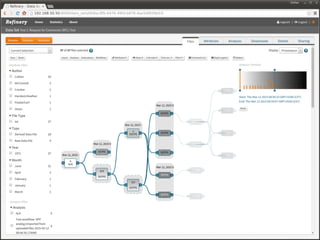
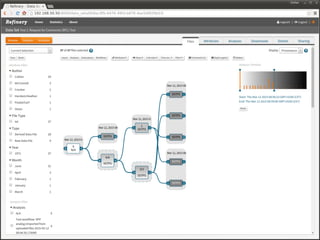
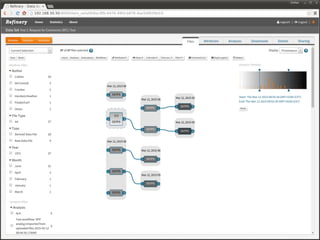
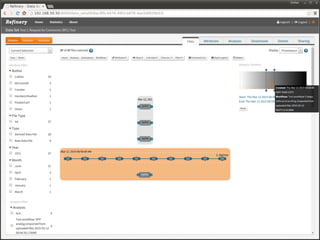
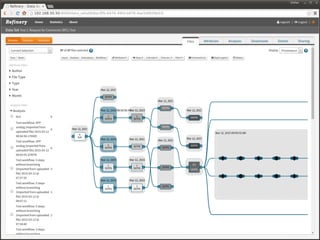
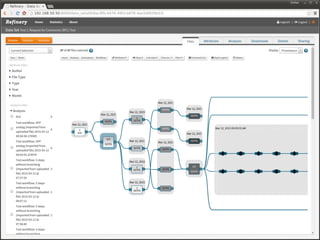
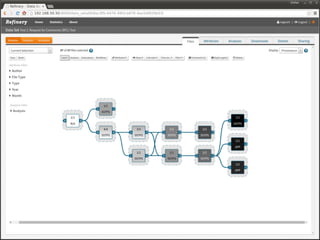
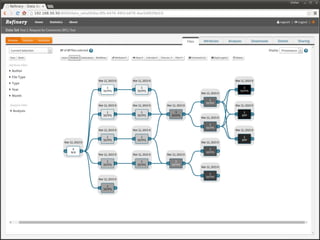
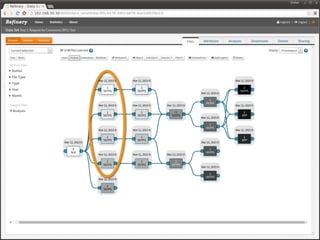
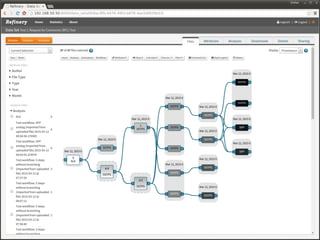
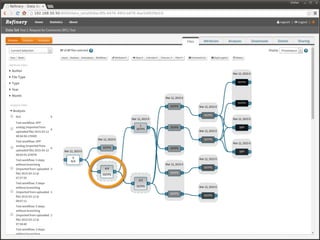
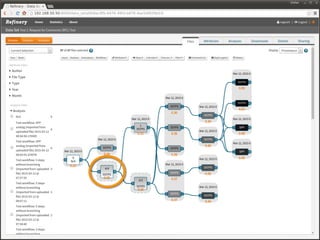
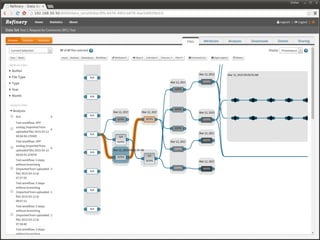
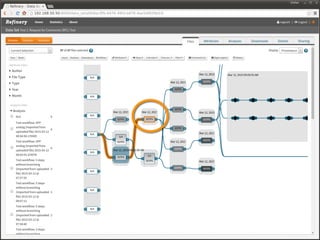
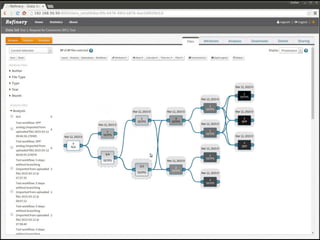
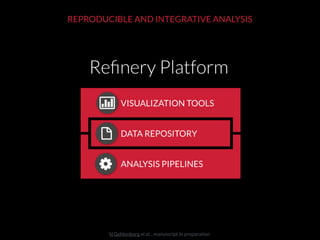
The document discusses issues related to reproducibility in scientific research, particularly highlighting the case of Diederik Stapel, who was found to have fabricated data. It emphasizes a growing movement among the scientific community and pharmaceutical companies to improve data transparency and experimental rigor in order to address the high failure rates in clinical trials and ensure the validity of research findings. A proposal for enhanced reproducibility through better tools and data management practices is also outlined.