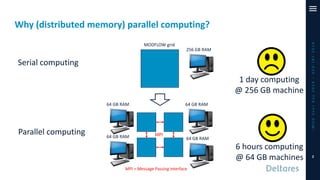
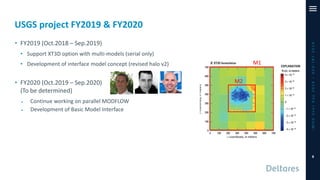
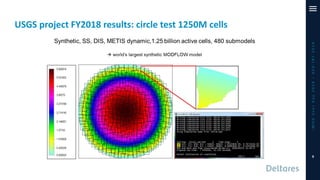
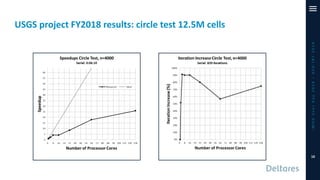
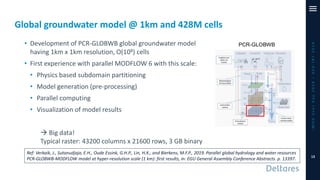
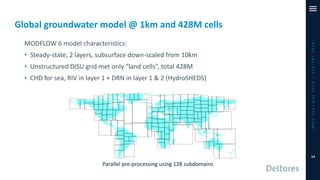
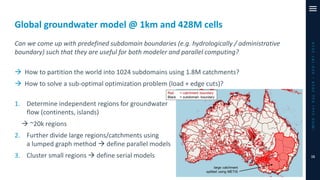
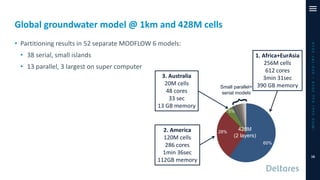
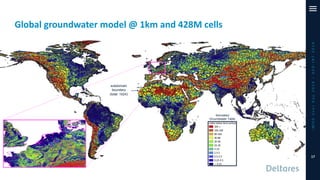
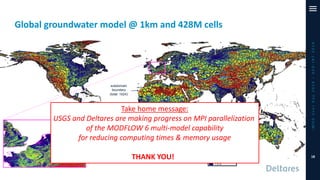
The document outlines a parallelization project for MODFLOW 6 undertaken by USGS and Deltares aimed at improving groundwater simulation through distributed memory parallel computing on high-performance supercomputers. Key components include the development of a parallel linear solver, the partitioning of large-scale models into subdomains, and the integration of new modeling capabilities for global groundwater applications. The project emphasizes reducing computing time and memory usage, with various results and ongoing plans for future development presented.