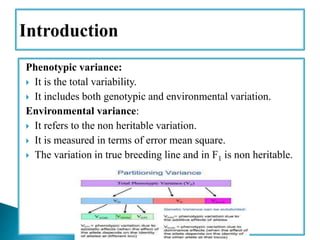
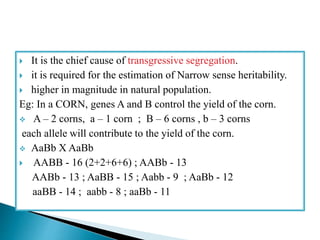
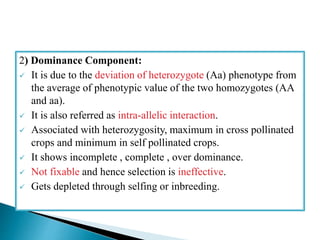
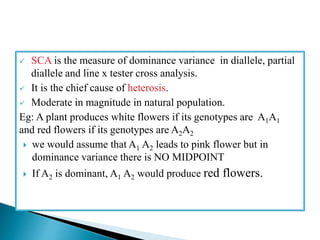
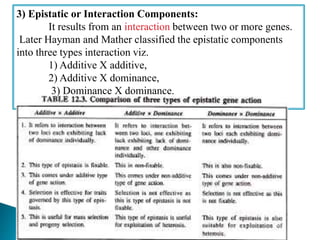
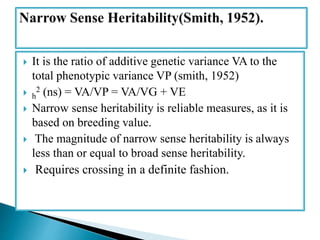
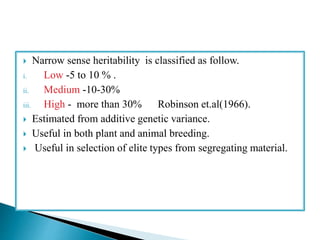
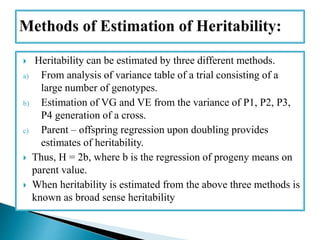
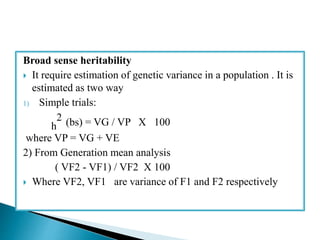
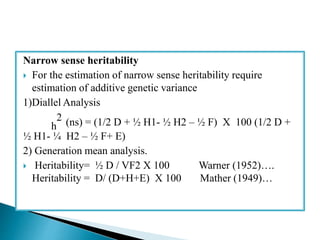
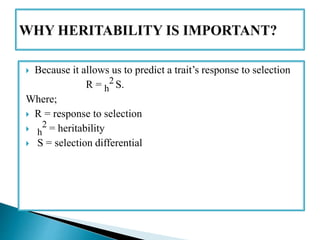
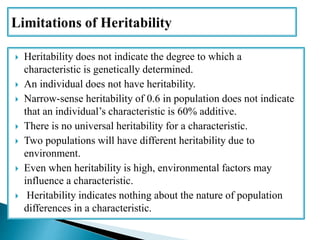
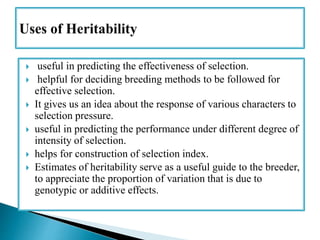
This document discusses the key concepts of phenotypic variance, environmental variance, genetic variance, heritability, and the different components of genetic variance. It explains that phenotypic variance is the total observable variability and includes both genotypic and environmental influences. Genetic variance refers only to the inheritable portion that is important for crop improvement. Genetic variance can be further divided into additive, dominance, and epistatic components. The document also defines broad and narrow sense heritability and how they are estimated. It provides examples to illustrate concepts like additive and dominance effects.