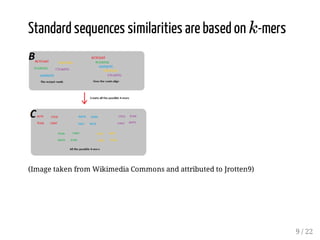
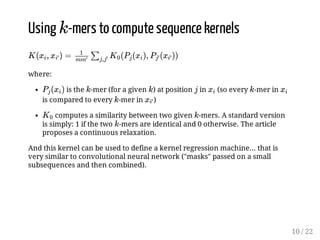
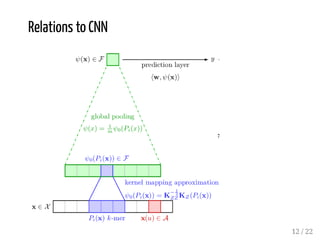
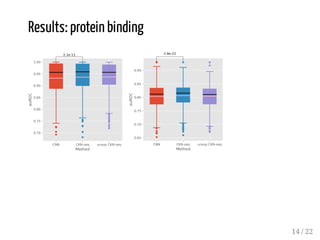
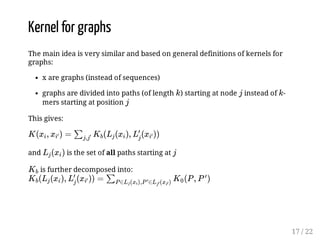
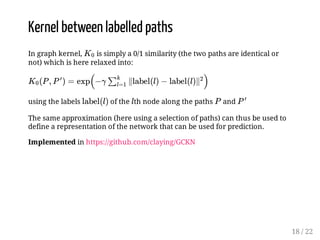
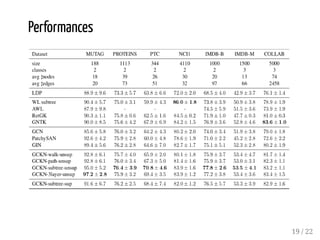
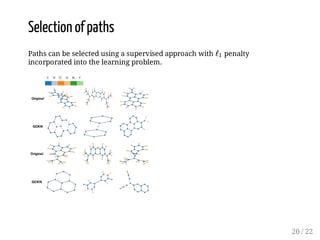
This presentation discusses how convolutional kernel networks (CKNs) can be used to model sequential and graph-structured data through kernels defined over sequences and graphs. CKNs define feature maps from substructures like n-mers in sequences and paths in graphs into high-dimensional spaces, which are then approximated to obtain low-dimensional representations that can be used for prediction tasks like classification. This approach is analogous to convolutional neural networks and can be extended to multiple layers. The presentation provides examples showing CKNs achieve good performance on problems involving protein sequences and social networks.


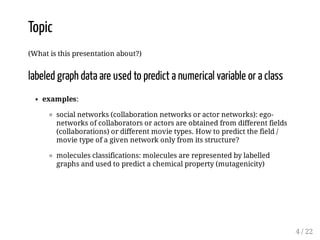
![Topic
(What is this presentation about?)
sequence data are used to predict a numerical variable or a class
sequences are vectors of dimension
examples:
protein homology: predicting the family of a protein from its
sequence
using DNA sequence to predict if the site is a TF binding site
[0, 1]
|A|×L
3 / 22](https://image.slidesharecdn.com/vialaneixwggnn2020-05-07-200925062134/85/Convolutional-networks-and-graph-networks-through-kernels-3-320.jpg)