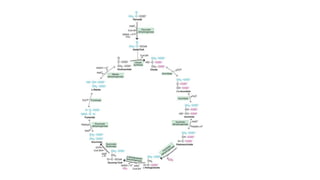
The citric acid cycle is the key metabolic pathway that generates energy in the form of ATP and NADH through the oxidation of acetyl-CoA derived from carbohydrates, fats, and proteins. The cycle's 10 steps occur in the mitochondrial matrix and involve enzymatic conversions between citrate, isocitrate, alpha-ketoglutarate, succinyl-CoA, succinate, fumarate, malate, and oxaloacetate. Two carbon atoms are removed as CO2 from acetyl-CoA in each turn of the cycle, generating high-energy electron carriers used in oxidative phosphorylation to produce ATP.