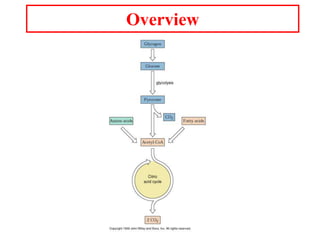
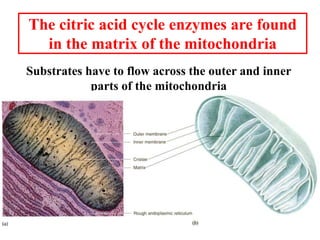
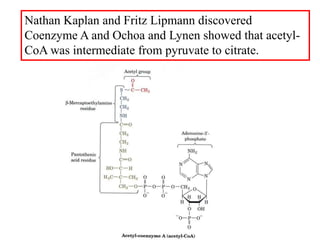
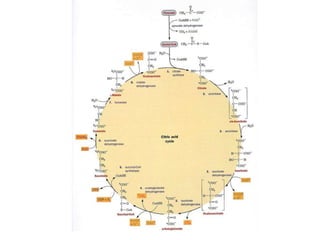
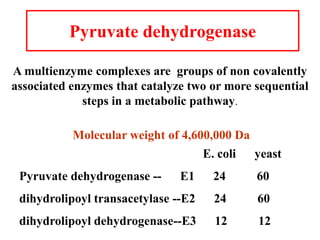
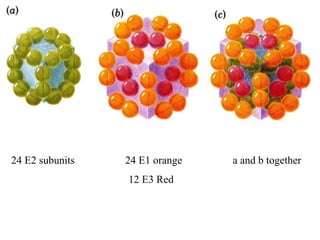
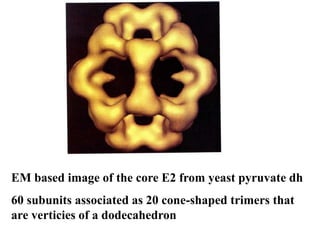
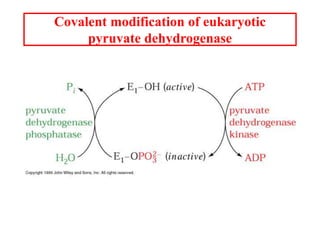
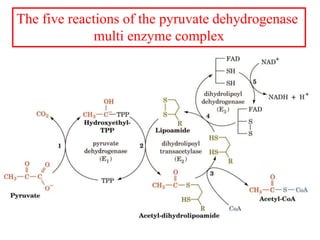
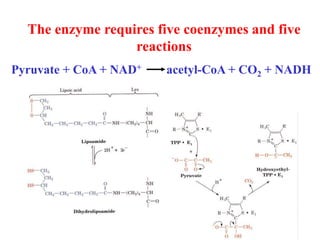
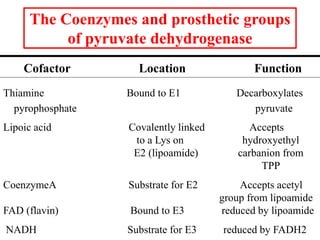
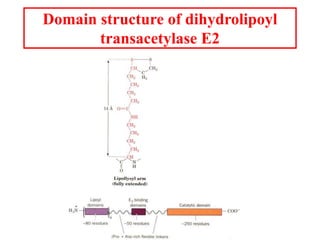
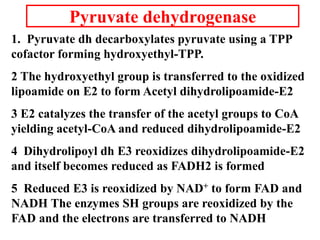
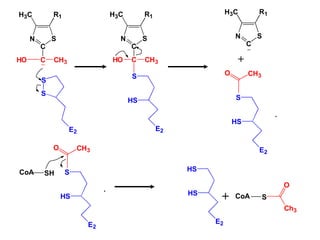
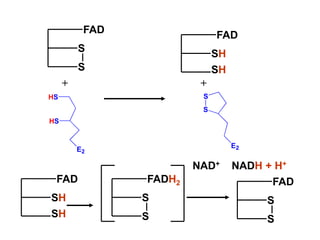
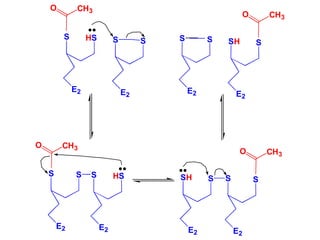
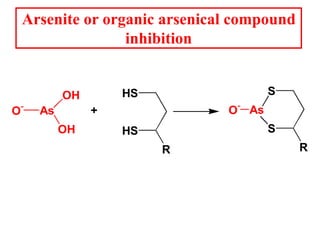
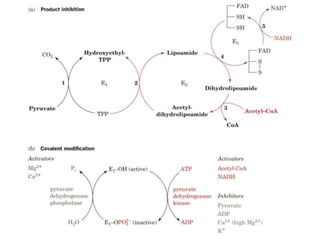
The citric acid cycle (TCA cycle) is a central metabolic pathway that catalyzes the oxidation of acetyl-CoA derived from carbohydrates, fats, and proteins, producing carbon dioxide and reducing equivalents in the form of NADH and FADH2 that are used in oxidative phosphorylation. It was established in the 1930s that TCA cycle intermediates stimulated oxygen consumption and CO2 release. Key discoveries in the 1940s-1950s showed that acetyl-CoA is the intermediate connecting pyruvate to the TCA cycle, and that the TCA cycle enzymes are organized in the mitochondrial matrix.