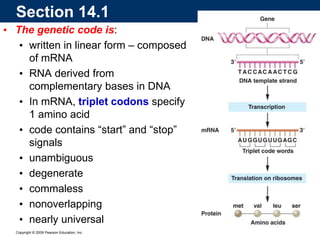
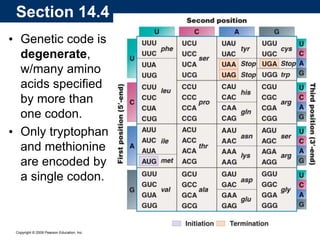
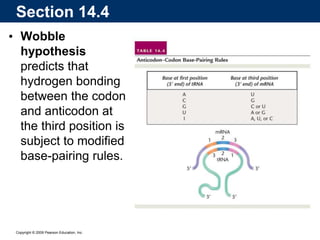
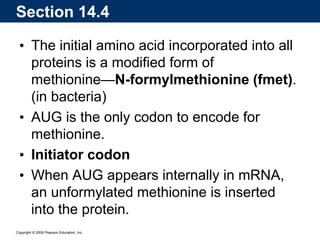
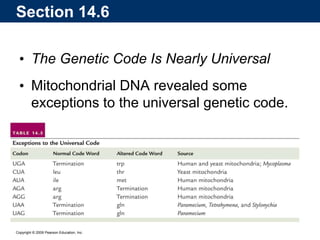
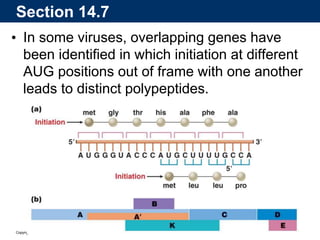
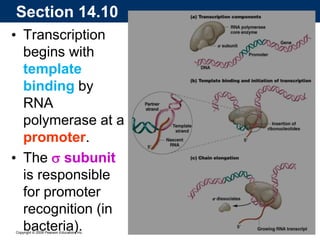
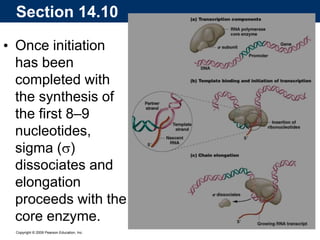
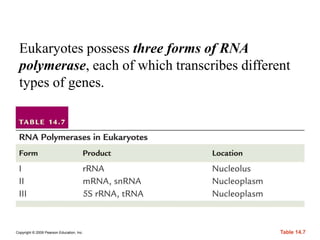
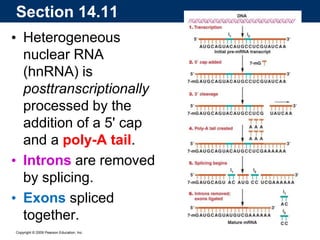
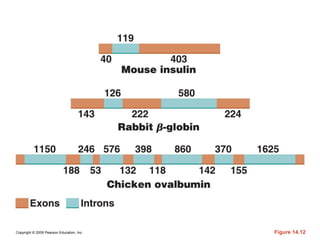
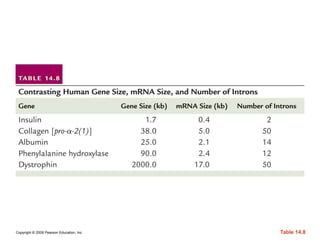
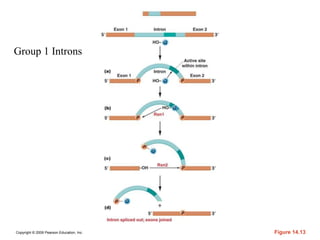
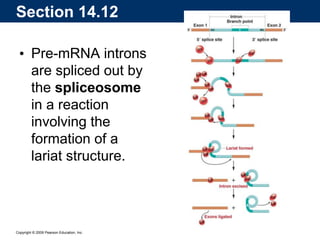
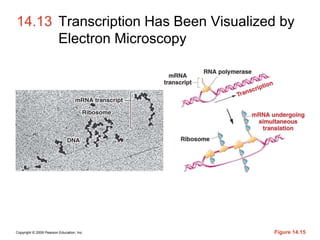
The document summarizes key aspects of the genetic code and transcription. It describes how the genetic code is written in mRNA using triplets of nucleotides that specify amino acids. It also explains that transcription in eukaryotes involves RNA polymerase II, promoters, and results in a pre-mRNA that undergoes splicing to remove introns and produce the mature mRNA. Visualization by electron microscopy has provided insights into the transcription process.