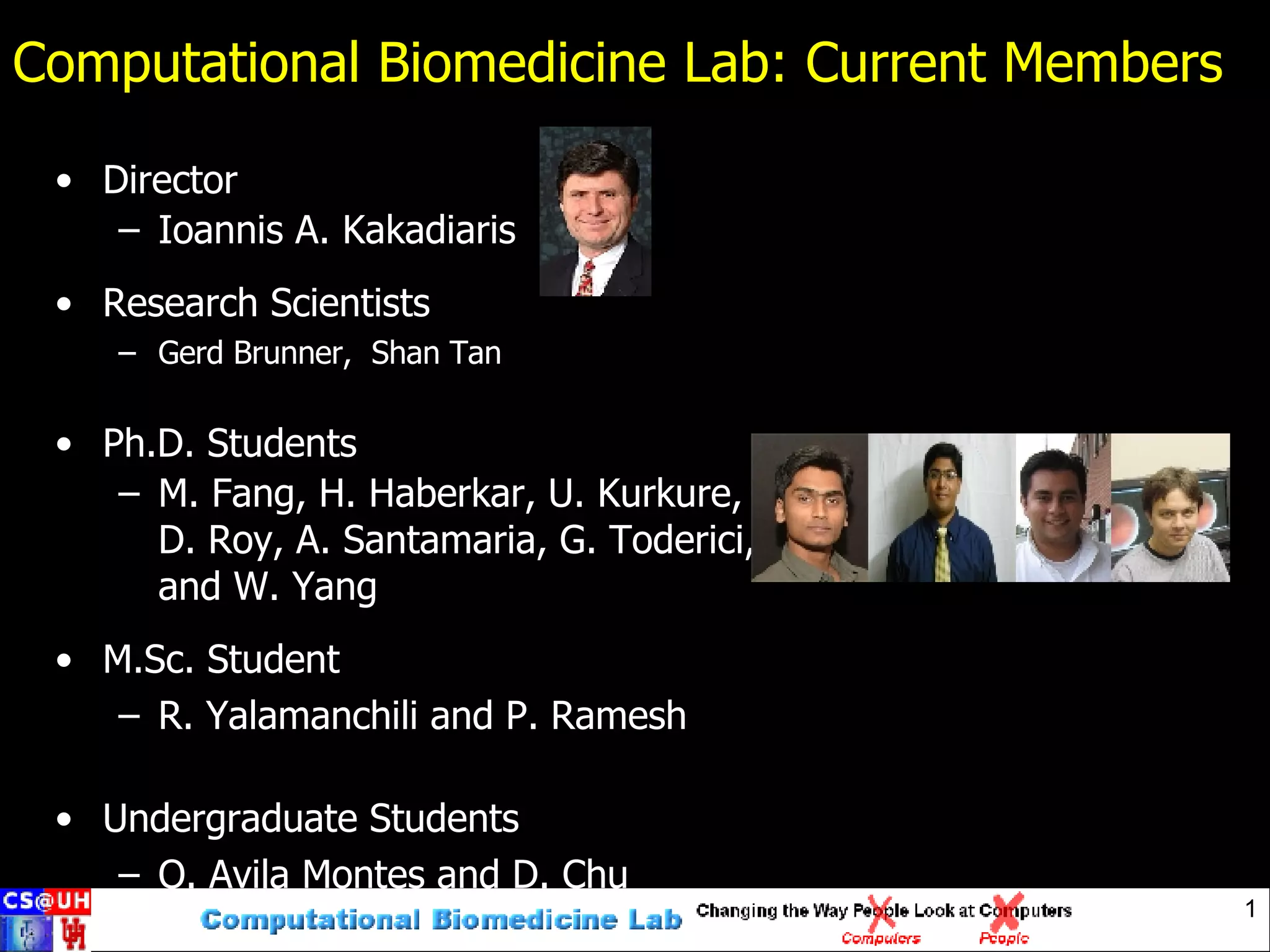
The Computational Biomedicine Lab has the following members: a director, research scientists, PhD students, an MSc student, and undergraduate students. The lab's mission is to develop algorithms to analyze multidimensional data and extract relevant information to aid humans. The lab aims to establish interdisciplinary collaboration and work on "grand challenge problems".

![CBL Roadmap New Computational Tools For Scientific Discovery From Algorithm to Bedside / TestBed Research Teams of the Future [email_address]](https://image.slidesharecdn.com/cbl-pp1-1195577899809833-4/75/Computational-Biomedicine-Lab-Current-Members-pumpsandpipesmdhc-3-2048.jpg)

![CS@UH research highlights: people’s hearts and minds [email_address] People’s hearts and minds](https://image.slidesharecdn.com/cbl-pp1-1195577899809833-4/75/Computational-Biomedicine-Lab-Current-Members-pumpsandpipesmdhc-6-2048.jpg)





![Roadmap New Computational Tools For Scientific Discovery From Algorithm to Bedside / TestBed Research Teams of the Future [email_address]](https://image.slidesharecdn.com/cbl-pp1-1195577899809833-4/75/Computational-Biomedicine-Lab-Current-Members-pumpsandpipesmdhc-25-2048.jpg)

![Contact Us Computational Biomedicine Lab http://www.cbl.uh.edu/ Prof. Ioannis A. Kakadiaris http://www.cbl.uh.edu/~ioannisk [email_address]](https://image.slidesharecdn.com/cbl-pp1-1195577899809833-4/75/Computational-Biomedicine-Lab-Current-Members-pumpsandpipesmdhc-27-2048.jpg)