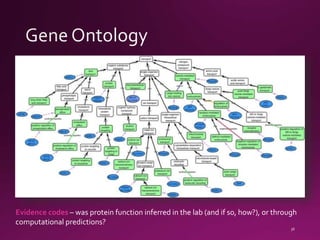
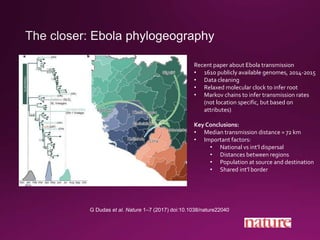
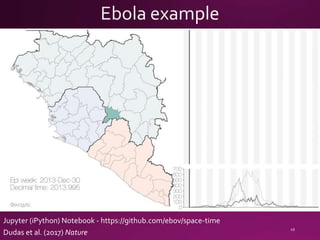
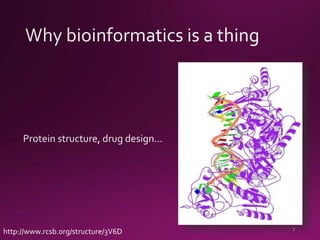
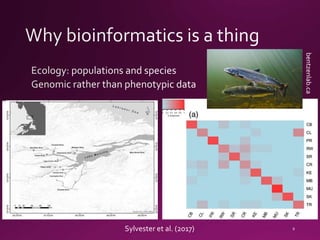
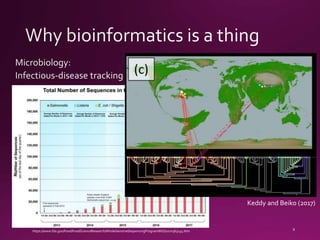
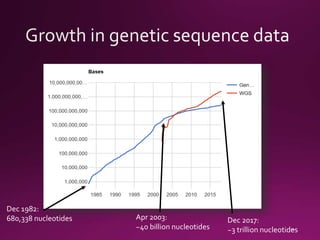
This document contains a summary of various topics related to bioinformatics and genomics including references to databases, tools, standards, and publications. It discusses databases of gene and protein sequences from as early as the 1960s to the present with trillions of nucleotides currently archived. Formats for sharing quantitative and gene expression data are mentioned as well as the importance of recording experiment details to allow comparisons and replication. Overall, the document provides an overview of the history and current state of genomics data sharing and analysis.





















![All articles published in Science in 2009 that mention H1N1:
https://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi?db=pubmed&term=science[
journal]+AND+h1n1+AND+2009[pdat]](https://image.slidesharecdn.com/beiko-biomedical-2019-190322131956/85/Biomedical-data-32-320.jpg)