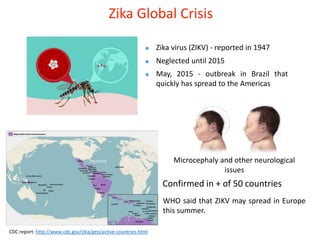
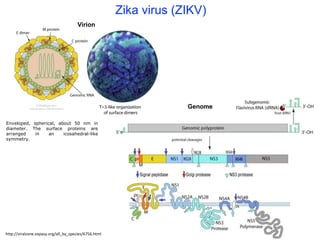
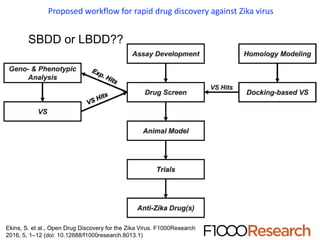
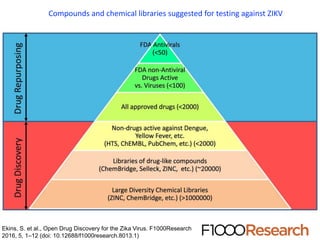
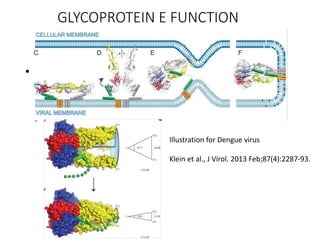
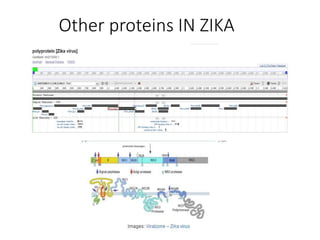
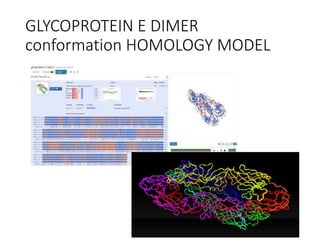
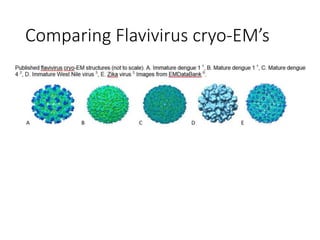
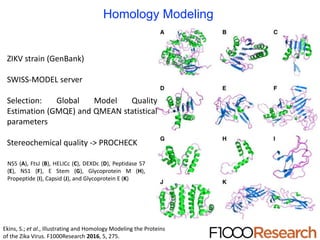
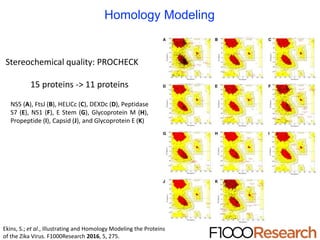
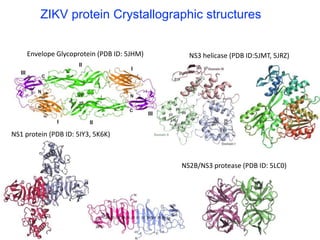
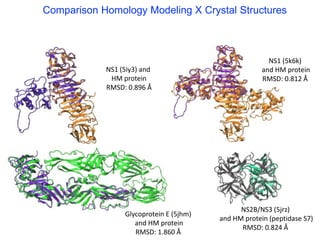
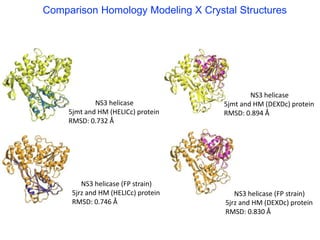
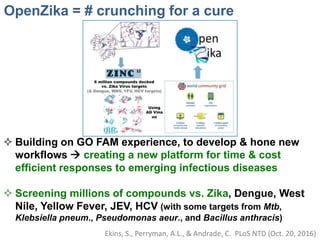
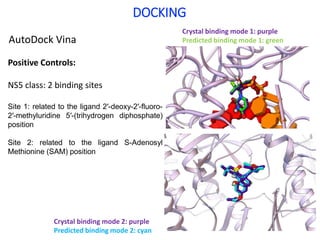
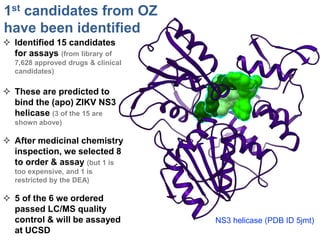
The document outlines the collaborative initiative 'OpenZika', which aims to accelerate drug discovery for the Zika virus through crowd-sourced computing power and various scientific collaborations. It details discussions and actions taken by researchers, including the use of open repositories and partnerships to address the public health crisis posed by Zika, particularly related to its neurological effects. It also highlights the progress in identifying potential antiviral drug candidates through extensive virtual screening of chemical compounds.