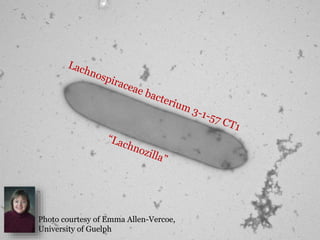
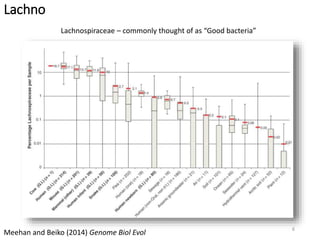
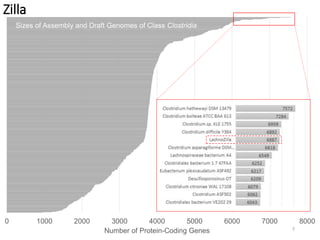
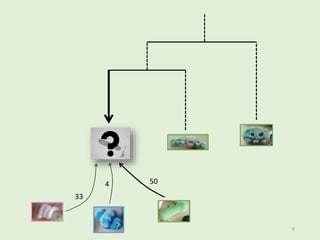
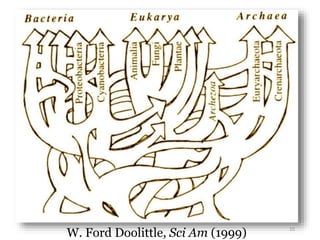
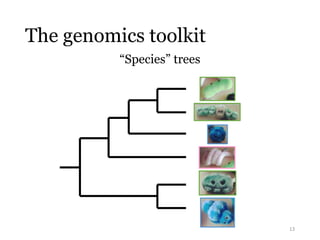
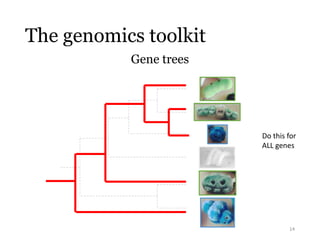
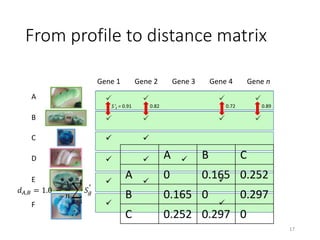
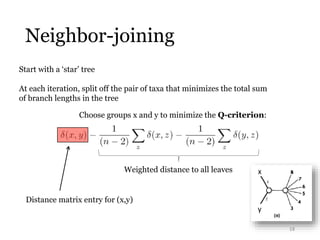
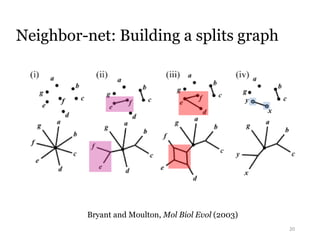
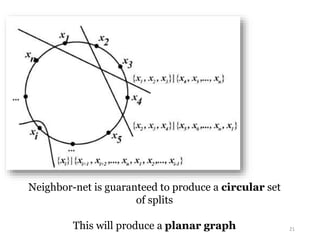
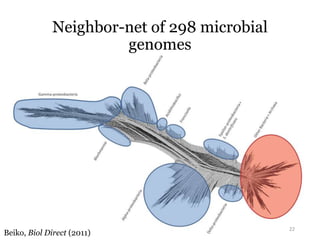
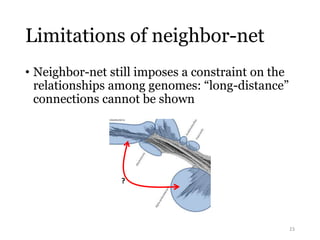
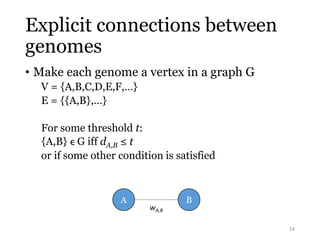
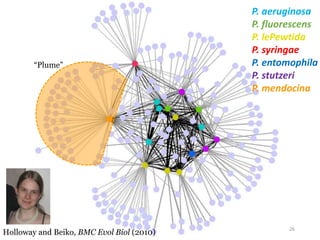
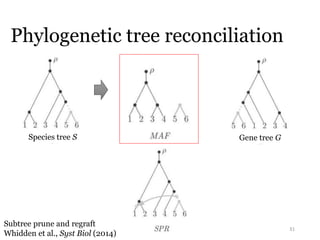
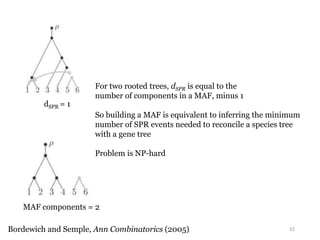
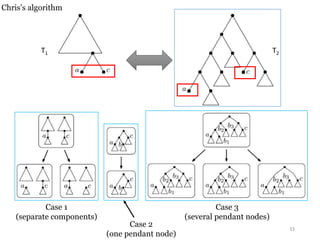
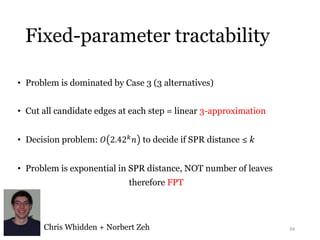
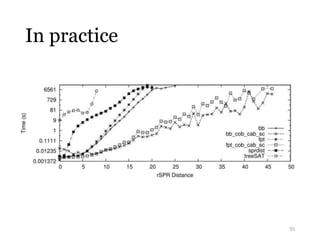
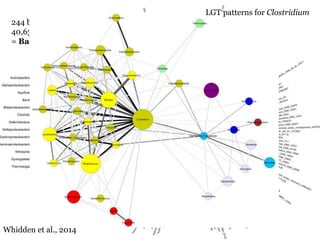
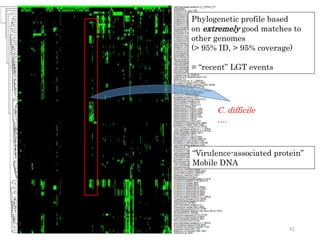
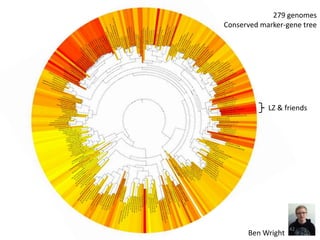
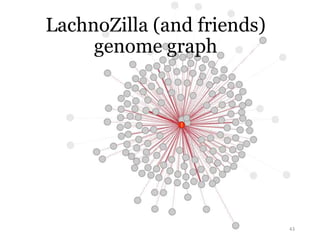
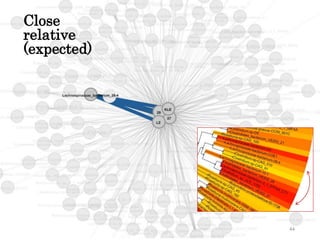
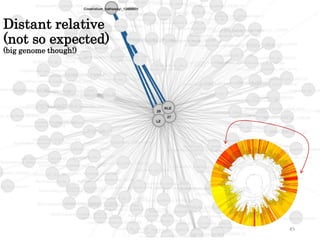
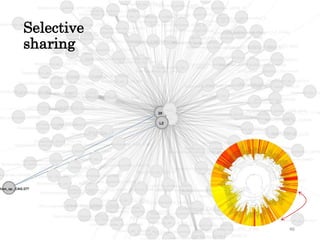
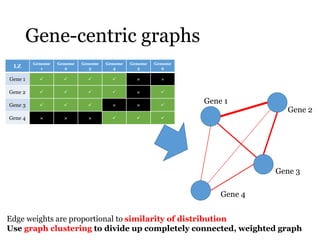
This document summarizes research on analyzing microbial communities and relationships using genomic tools. It discusses how gene profiles and trees can be used to understand microbial identities and relationships. Network approaches are presented as useful for capturing complex relationships between microbes, including lateral gene transfer. Phylogenetic reconciliation and supertree methods are described for inferring evolutionary histories and minimum gene transfer events needed to explain gene tree discordance with species trees. The analysis of one particular microbe, Lachnozilla, is discussed as a case study.